Exhibit 99.1

Enumeral Drug Discovery & Development July 2015 Program Updates

2 Overview • Program Overview • PD - 1 • TIM - 3 • TCR recovery (single cell) 2

Program Overview
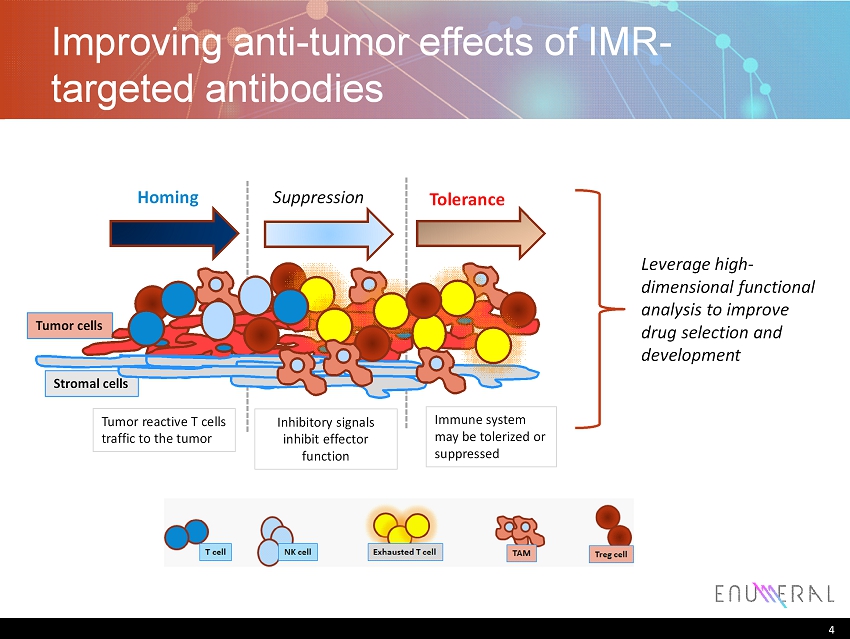
4 Improving anti - tumor effects of IMR - targeted antibodies 4 Tumor cells Stromal cells Homing Suppression Tolerance Tumor reactive T cells traffic to the tumor Inhibitory signals inhibit effector function Immune system may be tolerized or suppressed Leverage high - dimensional functional analysis to improve drug selection and development
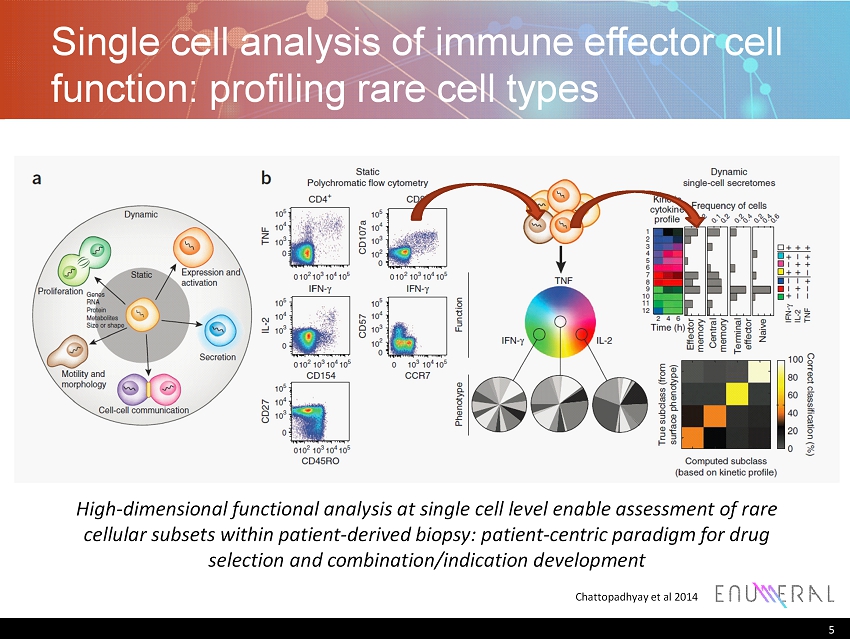
Single cell analysis of immune effector cell function: profiling rare cell types 5 Chattopadhyay et al 2014 High - dimensional functional analysis at single cell level enable assessment of rare cellular subsets within patient - derived biopsy: patient - centric paradigm for drug selection and combination/indication development
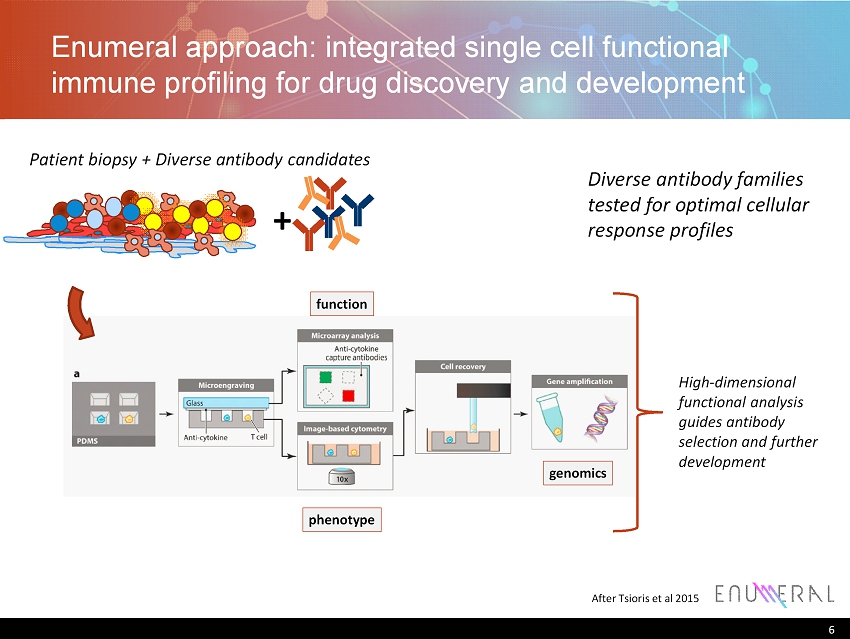
Enumeral approach: integrated single cell functional immune profiling for drug discovery and development 6 After Tsioris et al 2015 function phenotype genomics Patient biopsy + Diverse antibody candidates High - dimensional functional analysis guides antibody selection and further development Diverse antibody families tested for optimal cellular response profiles +
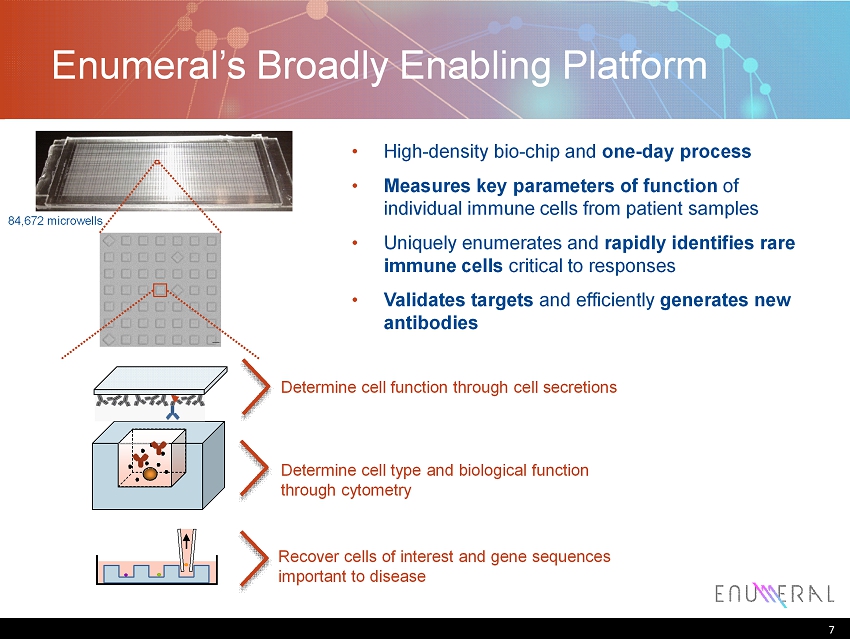
7 Enumeral’s B roadly E nabling P latform 7 84,672 microwells • High - density bio - chip and one - day process • Measures key parameters of function of individual immune cells from patient samples • Uniquely enumerates and rapidly identifies rare immune cells critical to responses • Validates targets and efficiently generates new antibodies Determine cell function through cell secretions Determine cell type and biological function through cytometry R ecover cells of interest and gene sequences important to disease

ENUM003: PD - 1 8
9 Background: screening design • Two strains of mice were immunized with PD - 1 protein with multiple types of adjuvant • Serum titres were extremely high • Prior to screening, B cell activation was assessed (BLIMP1+) • Screening was performed using primary B cells, specifically screening secreted antibodies for: – Isotype: IgM vs IgG – PD - 1 – Lag3 • PD1+ IgG+ microarray events were gated for isolation and recovery of B cells for RT - PCR 9
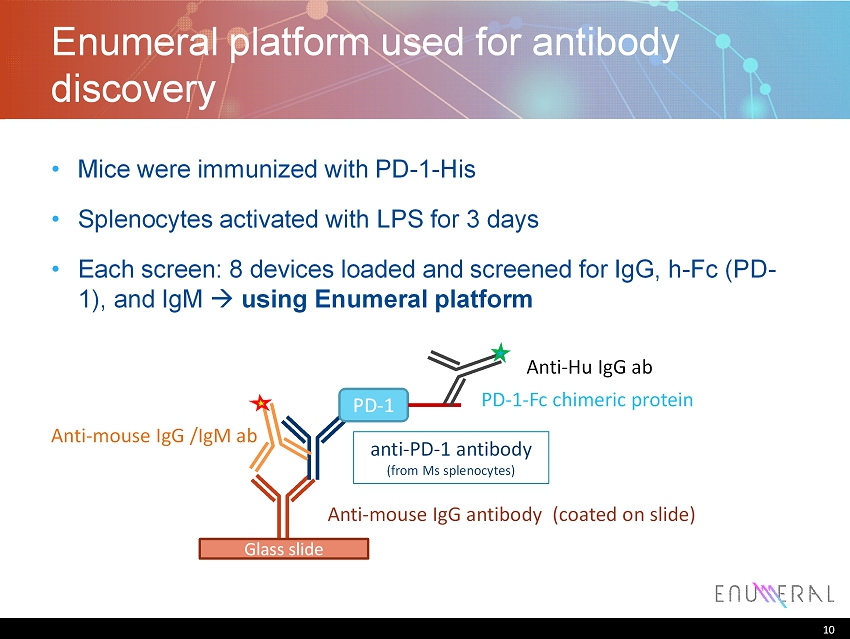
10 Enumeral platform used for antibody discovery • Mice were immunized with PD - 1 - His • Splenocytes activated with LPS for 3 days • Each screen: 8 devices loaded and screened for IgG, h - Fc (PD - 1), and IgM using Enumeral platform 10 Glass slide Anti - mouse IgG antibody (coated on slide) anti - PD - 1 antibody (from Ms splenocytes ) PD - 1 PD - 1 - Fc chimeric protein Anti - mouse IgG /IgM ab Anti - Hu IgG ab
11 Raw Hit Rate/Lead Discovery • >500 IgG+ PD - 1+ raw screening hits • On average: – Recovery rate of 57.6% matched pairs per screen – 84.2% of those matched pairs showed binding to rhPD - 1 11
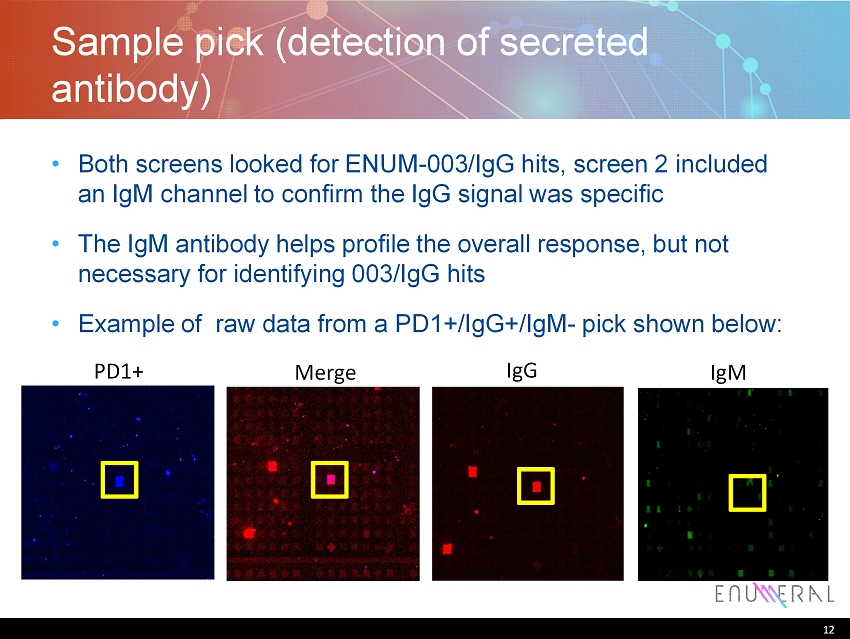
12 Sample pick (detection of secreted antibody) • Both screens looked for ENUM - 003/IgG hits, screen 2 included an IgM channel to confirm the IgG signal was specific • The IgM antibody helps profile the overall response, but not necessary for identifying 003/IgG hits • Example of raw data from a PD1+/IgG+/IgM - pick shown below: 12 PD1+ Merge IgG IgM
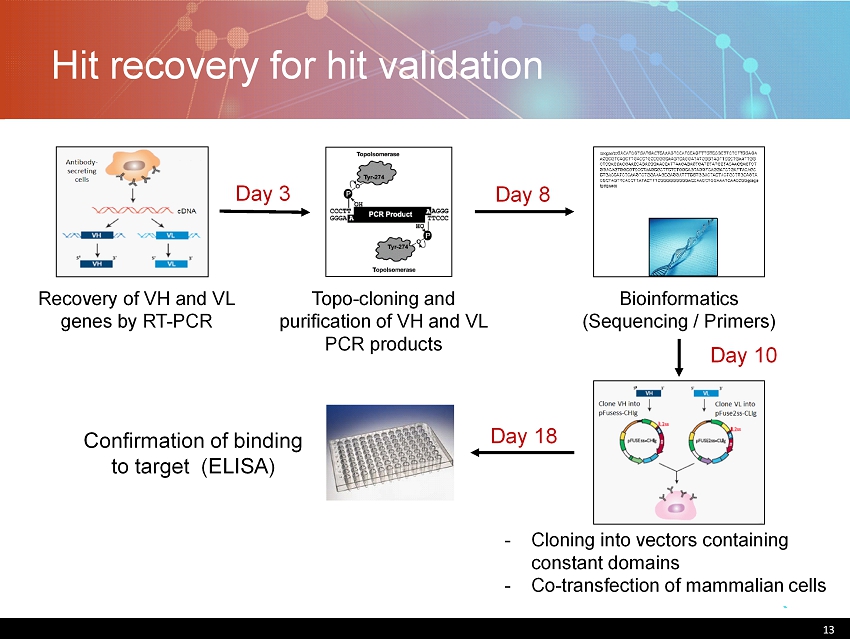
Hit recovery for hit validation 13 Bioinformatics (Sequencing / Primers) Recovery of VH and VL genes by RT - PCR Topo - cloning and purification of VH and VL PCR products Confirmation of binding to target (ELISA) Day 3 Day 8 Day 10 Day 18 - Cloning into vectors containing constant domains - Co - transfection of mammalian cells
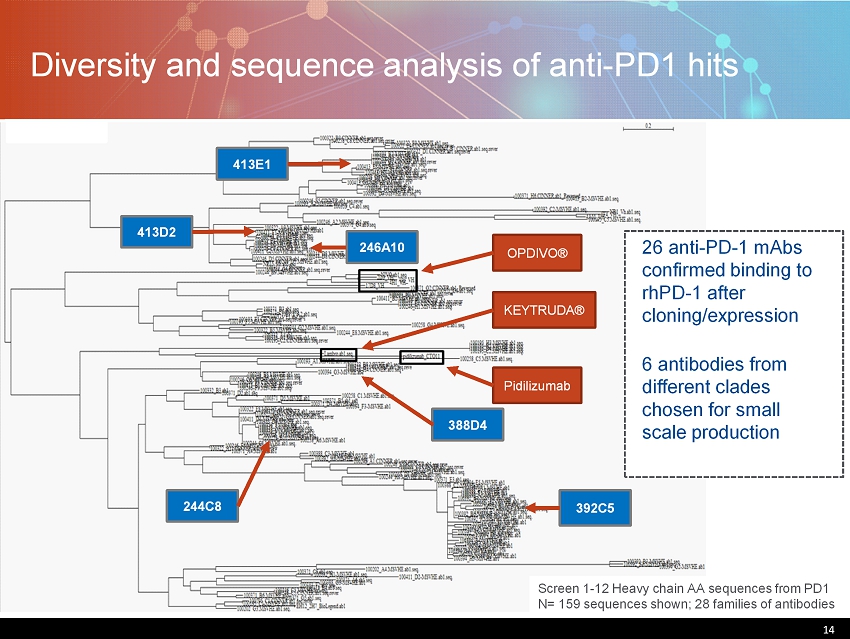
14 Diversity and sequence analysis of anti - PD1 hits 14 Screen 1 - 12 Heavy chain AA sequences from PD1 N= 159 sequences shown; 28 families of antibodies OPDIVO® KEYTRUDA® Pidilizumab 26 anti - PD - 1 mAbs confirmed binding to rhPD - 1 after cloning/expression 6 antibodies from different clades chosen for small scale production 392C5 246A10 413D2 413E1 244C8 388D4
15 Hit confirmation • n=26 clones were transiently transfected and cultured supernatants were used for hit confirmation testing • Binding confirmation assays: – ELISA: recombinant human PD1 and cynomolgus PD1 – HEK293 - PD1 cells – Primary T cells 15
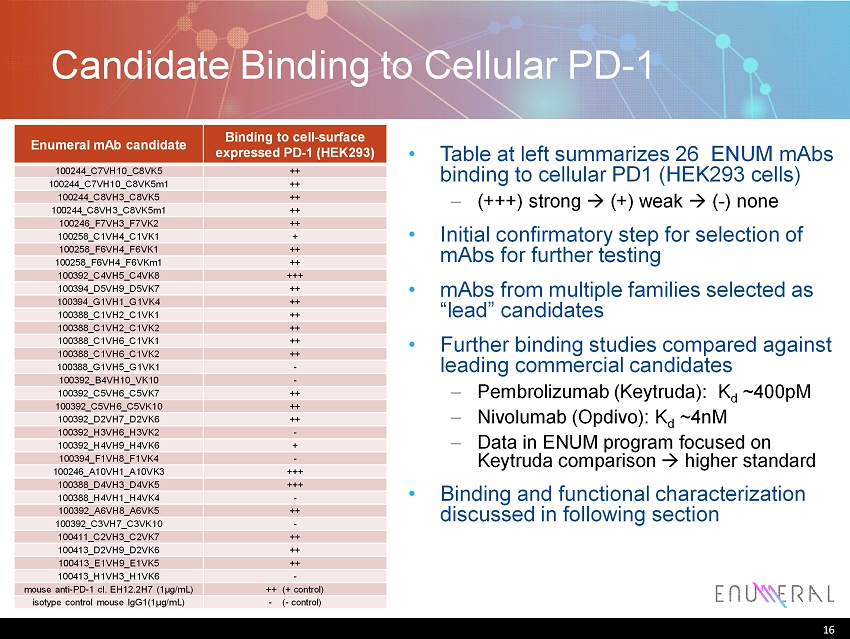
16 Enumeral mAb candidate Binding to cell - surface expressed PD - 1 (HEK293) 100244_C7VH10_C8VK5 ++ 100244_C7VH10_C8VK5m1 ++ 100244_C8VH3_C8VK5 ++ 100244_C8VH3_C8VK5m1 ++ 100246_F7VH3_F7VK2 ++ 100258_C1VH4_C1VK1 + 100258_F6VH4_F6VK1 ++ 100258_F6VH4_F6VKm1 ++ 100392_C4VH5_C4VK8 +++ 100394_D5VH9_D5VK7 ++ 100394_G1VH1_G1VK4 ++ 100388_C1VH2_C1VK1 ++ 100388_C1VH2_C1VK2 ++ 100388_C1VH6_C1VK1 ++ 100388_C1VH6_C1VK2 ++ 100388_G1VH5_G1VK1 - 100392_B4VH10_VK10 - 100392_C5VH6_C5VK7 ++ 100392_C5VH6_C5VK10 ++ 100392_D2VH7_D2VK6 ++ 100392_H3VH6_H3VK2 - 100392_H4VH9_H4VK6 + 100394_F1VH8_F1VK4 - 100246_A10VH1_A10VK3 +++ 100388_D4VH3_D4VK5 +++ 100388_H4VH1_H4VK4 - 100392_A6VH8_A6VK5 ++ 100392_C3VH7_C3VK10 - 100411_C2VH3_C2VK7 ++ 100413_D2VH9_D2VK6 ++ 100413_E1VH9_E1VK5 ++ 100413_H1VH3_H1VK6 - mouse anti - PD - 1 cl. EH12.2H7 (1μg/mL) ++ (+ control) isotype control mouse IgG1(1μg/mL) - ( - control) Candidate Binding to Cellular PD - 1 16 • Table at left summarizes 26 ENUM mAbs binding to cellular PD1 (HEK293 cells) – (+++) strong (+) weak ( - ) none • Initial confirmatory step for selection of mAbs for further testing • mAbs from multiple families selected as “lead” candidates • Further binding studies compared against leading commercial candidates – Pembrolizumab ( Keytruda ): K d ~400pM – Nivolumab ( Opdivo ): K d ~ 4nM – Data in ENUM program focused on Keytruda comparison higher standard • Binding and functional characterization discussed in following section

17 Scale - up of lead clones • n=6 lead clones were selected for further characterization • ELISA, EC50 determination • SPR, biophysical determination • Binding to cells, EC50 (MFI) • Mapping epitope, using scanning overlapping peptides • Cross - competition, compared to known antibodies 17

18 Binding Affinity: ENUM mAbs vs. Keytruda mAb EC 50 Keytruda 0.066 ENUM - A10 0.020 ENUM - C8 0.005 ENUM - D4 0.003 ENUM - E1 0.013 ENUM - D2 0.005 18 0 0.5 1 1.5 2 2.5 3 3.5 OD (450nm) Antibody concentration ( μ g/mL)
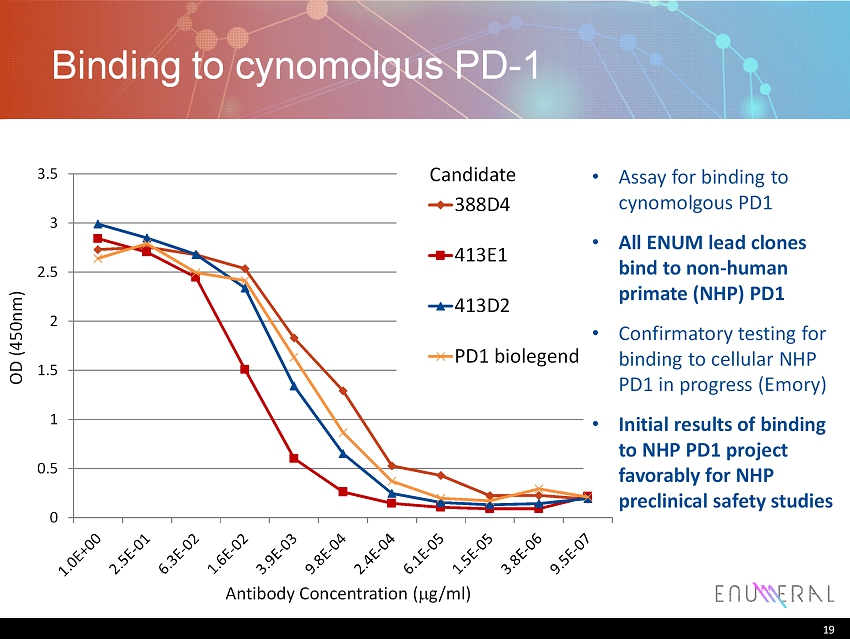
19 Binding to cynomolgus PD - 1 19 Antibody Concentration ( m g/ml) OD (450nm) 0 0.5 1 1.5 2 2.5 3 3.5 388D4 413E1 413D2 PD1 biolegend • Assay for binding to cynomolgous PD1 • All ENUM lead clones bind to non - human primate (NHP) PD1 • Confirmatory testing for binding to cellular NHP PD1 in progress (Emory) • Initial results of binding to NHP PD1 project favorably for NHP preclinical safety studies Candidate

20 Functional Characterization • Cell based assays for readout on PD1 enhancement of T cell function (proliferation, effector cell function) • PBMCs with exogenous rhPD - L1 - Ig • PBMCs with suboptimal stimulation conditions • PBMC - based MLR reaction • PBMC - based antigen recall (CMV or EBV) 20
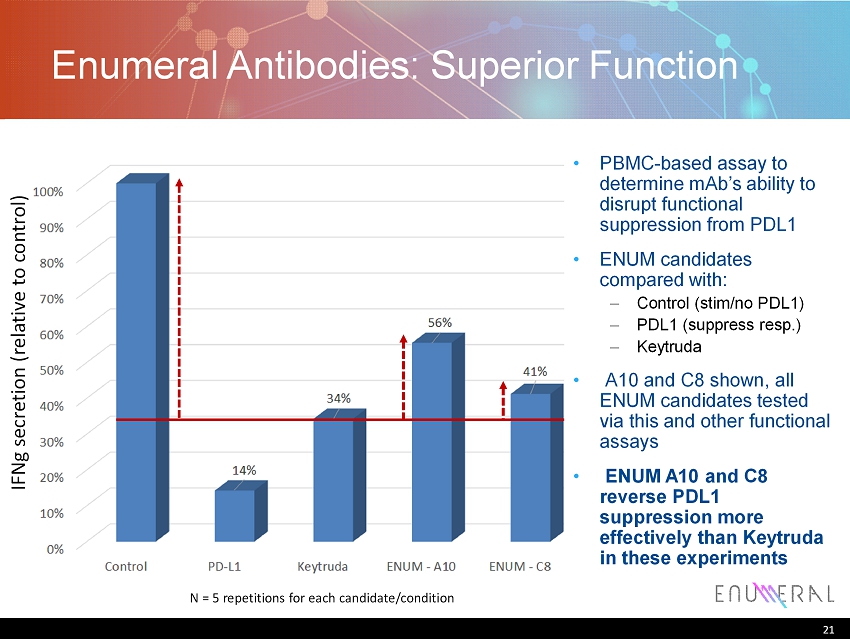
21 Enumeral Antibodies: Superior Function 21 IFNg secretion (relative to control) N = 5 repetitions for each candidate/condition • PBMC - based assay to determine mAb’s ability to disrupt functional suppression from PDL1 • ENUM candidates compared with : – Control ( stim /no PDL1) – PDL1 (suppress resp.) – Keytruda • A10 and C8 shown, all ENUM candidates tested via this and other functional assays • ENUM A10 and C8 reverse PDL1 suppression more effectively than Keytruda in these experiments
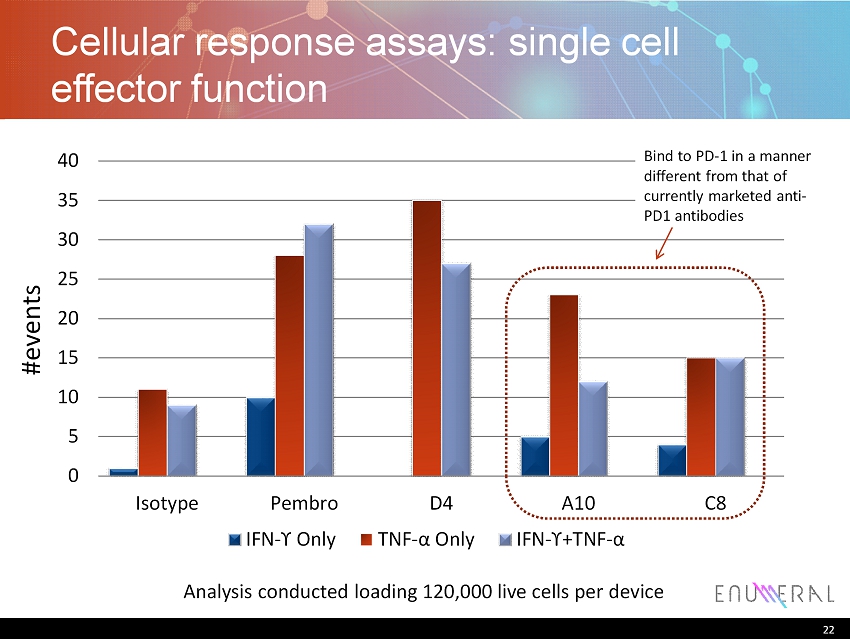
22 Cellular response assays: single cell effector function 0 5 10 15 20 25 30 35 40 Isotype Pembro D4 A10 C8 IFN - ϒ Only TNF - α Only IFN - ϒ+ TNF - α Analysis conducted loading 120,000 live cells per device #events 22 Bind to PD - 1 in a manner different from that of currently marketed anti - PD1 antibodies
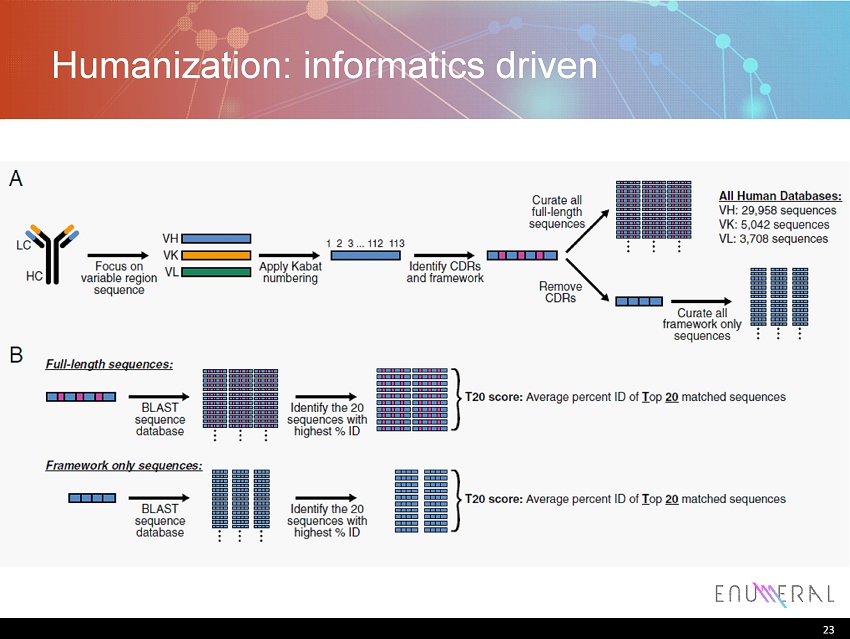
Humanization: informatics driven 23
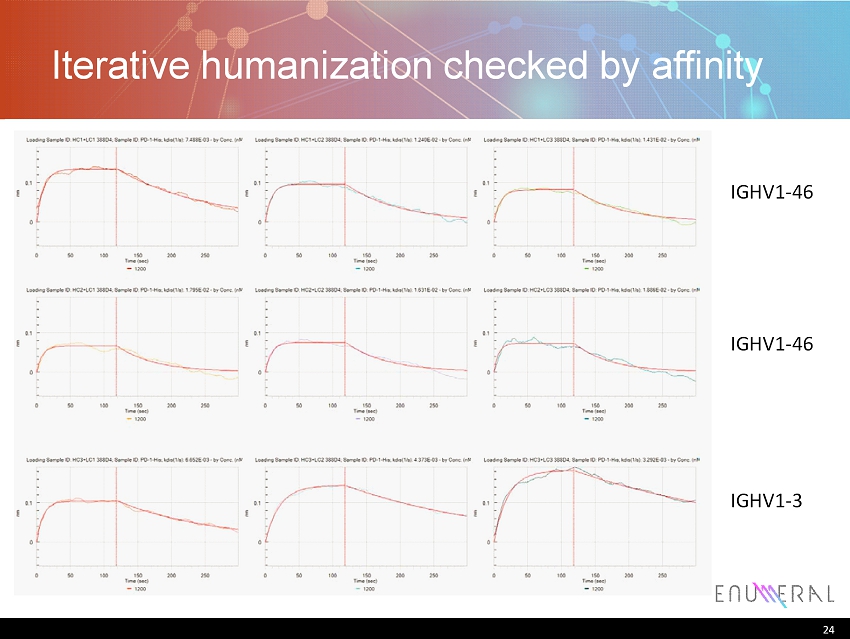
24 Iterative humanization checked by affinity 24 IGHV1 - 46 IGHV1 - 46 IGHV1 - 3

25 Humanization • n =4 clones selected for humanization – Humanization of 388D4 completed • In progress: – Binding: confirm binding and affinity – Specificity: confirm on - mechanism binding – Affinity: assessment compared to clinical standards – Safety: specificity against normal tissue panels 25

ENUM005: TIM - 3 26
27 TIM - 3 updates • 3 screens were processed (one BL6 and two NZB) • 169 cells were picked for RT - PCR • 88 matched pairs were recovered representing 30 unique antibodies • 8 reformatted antibodies were showed to bind rhTIM - 3 • 41 other antibodies are undergoing cloning/expression • More screens are planned to reach saturation 27
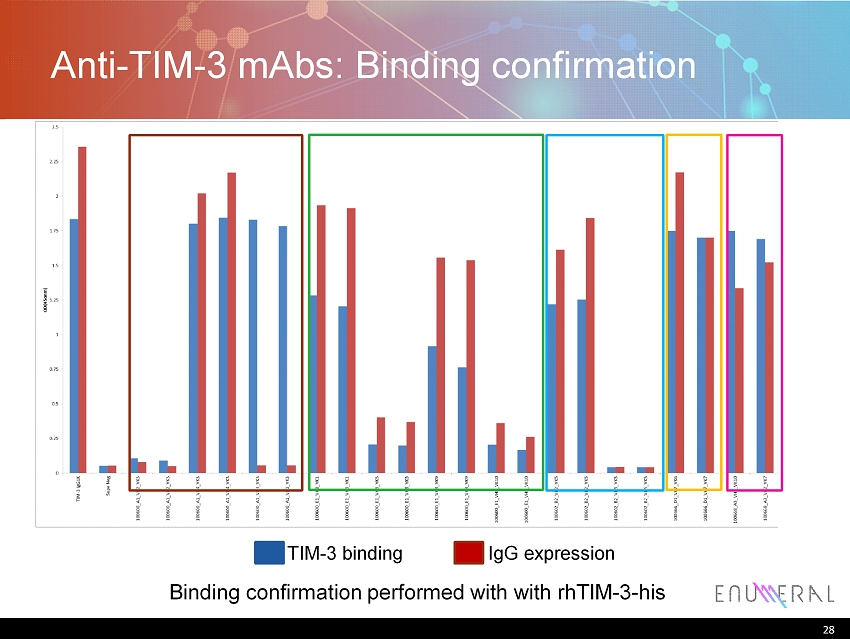
28 Anti - TIM - 3 mAbs : Binding confirmation 28 Binding confirmation performed with with rhTIM - 3 - his IgG expression TIM - 3 binding
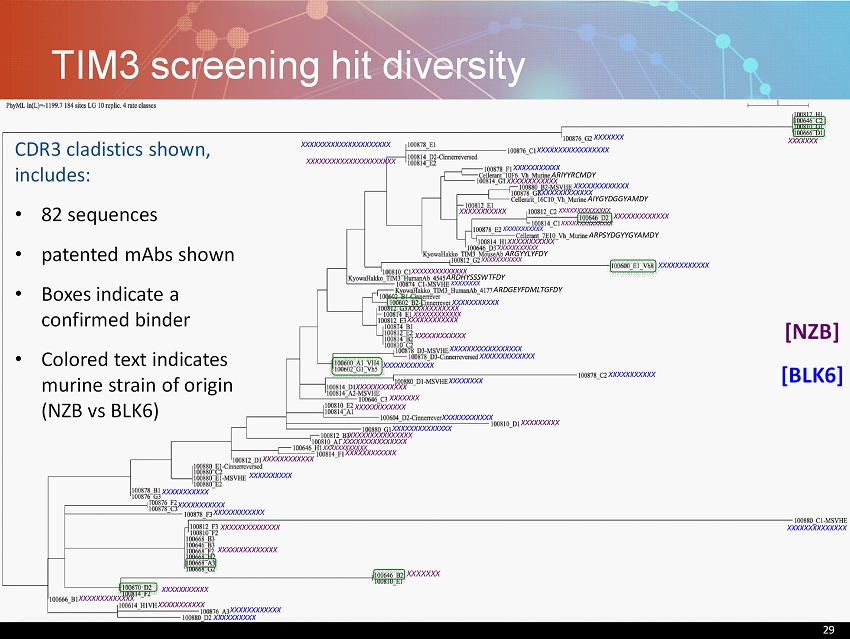
29 TIM3 screening hit diversity 29 [NZB] [BLK6] XXXXXXXXXXX XXXXXXXXXXXXXX XXXXXXXXXXXXXX AIYGYDGGYAMDY XXXXXXX XXXXXXX XXXXXXXXXXXX XXXXXXXXXXXXXXXXXXXXX XXXXXXXXXXXXXXXXXXXXX XXXXXXXXXXXXXXX XXXXXXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXXXXXX XXXXXXXXXXX ARIYYRCMDY ARGYYLYFDY XXXXXXXXXXX XXXXXXXXXXXX ARDHYSSSWTFDY ARPSYDGYYGYAMDY XXXXXXXXXXX XXXXXXXXXXX XXXXXXXXXXXX ARDGEYFDMLTGFDY XXXXXXXXX XXXXXXXXXXXXXX XXXXXXXXXXXXXX XXXXXXXXXXXXX XXXXXXXXXXXX XXXXXXX XXXXXXXXXXXXXXXXX XXXXXXXXXXX XXXXXXXXXXXXX XXXXXXXXXXXXX XXXXXXXXXXX XXXXXXXXXXX XXXXXXXXXXXXX XXXXXXXXXXXXXXXXX XXXXXXXX XXXXXXXXXXX XXXXXXXX XXXXXXX XXXXXXXXXXXX XXXXXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXX XXXXXXXXXXX XXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXXXXXX XXXXXXXXXXXXX XXXXXXXXXXX XXXXXXXXXXXX XXXXXXXXXX CDR3 cladistics shown, includes: • 82 sequences • patented mAbs shown • Boxes indicate a confirmed binder • Colored text indicates murine strain of origin (NZB vs BLK6)
30 Biochemical characterization of anti - TIM - 3 mAbs • Binding to rh - TIM - 3 - HIS by ELISA, and EC50 • Binding to cell - surface expressed TIM - 3 (HEK293 - TIM3+ cell line generated in house), and EC50 • Binding to human PBMCs • Binding to primate TIM - 3 protein and PBMCs • Epitope mapping • Affinity measurements 30

31 • Allo - MLR assay • Antigen recall assay (HIV or CMV) • Reversal of NK cell exhaustion (Melanoma) 31 Functional characterization of anti - TIM - 3 mAbs

32 TIM - 3 program timelines • More screening to increase diversity (saturation) – End of July 2015 • Biochemical characterization – Q4 ‘15 • Functional characterization – Q4 ‘15 • Humanization – Q4 ‘15 – Q1 ‘16 32

Human TCR recovery 33
34 Recovery of single T cells • Goal: use Enumeral platform to select specific T cells from patient samples and recover TCR gene sequences – Assess clonality of infiltrates – Assess repertoire changes/compare blood to disease microenvironment • Methods SOP development using Jurkat cells (following slides) 34
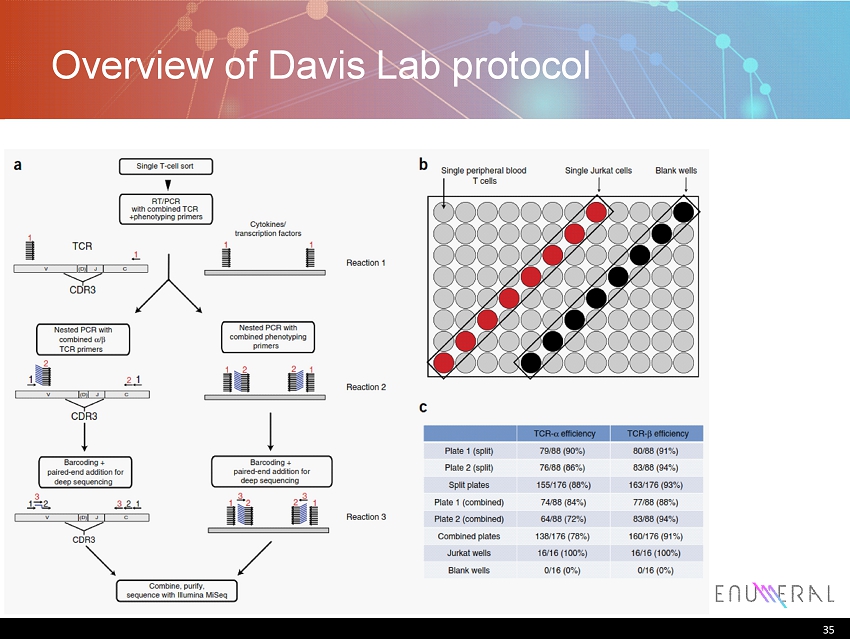
35 Overview of Davis Lab protocol 35
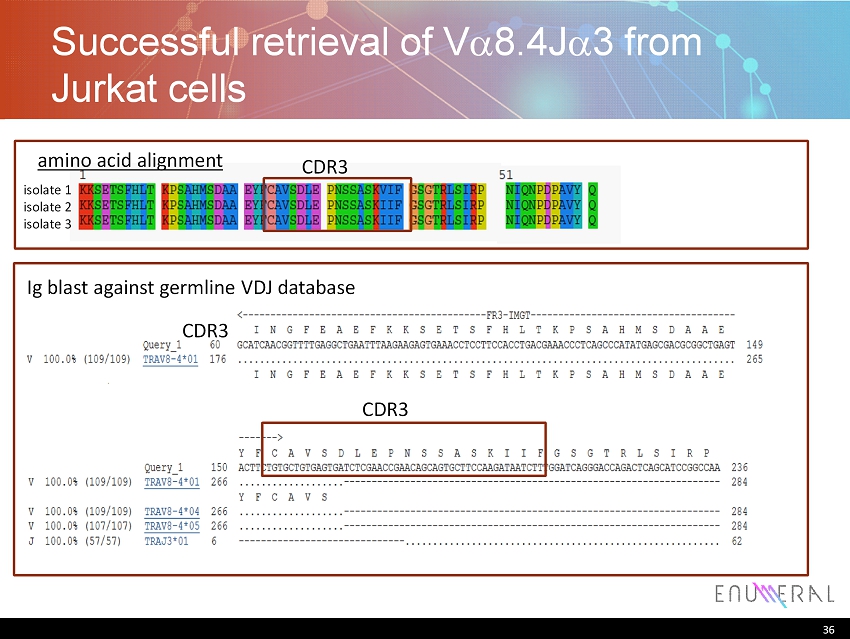
36 Successful retrieval of V a 8.4J a 3 from Jurkat cells 36 isolate 1 isolate 2 isolate 3 amino acid alignment Ig blast against germline VDJ database CDR3 CDR3 CDR3
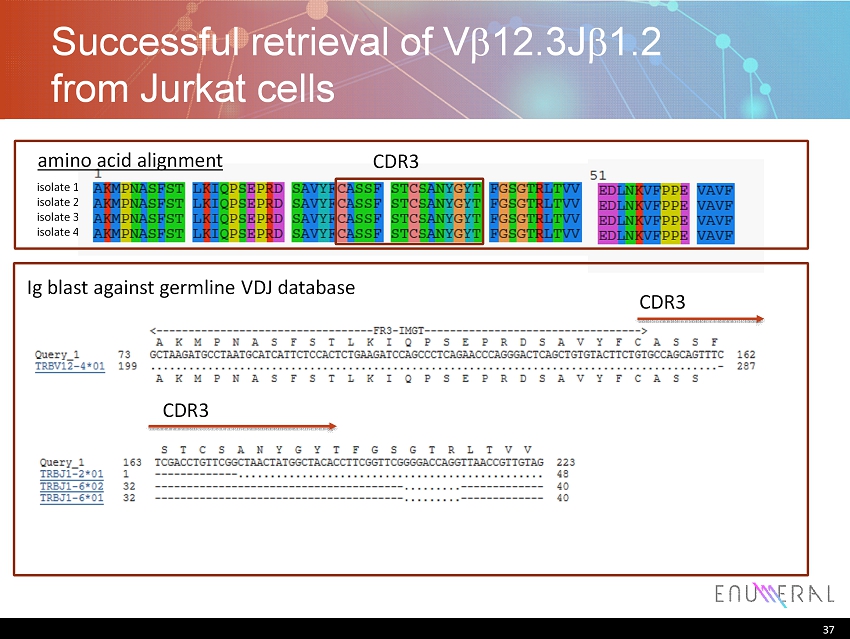
37 Successful retrieval of V b 12.3J b 1.2 from Jurkat cells 37 isolate 1 isolate 2 isolate 3 isolate 4 amino acid alignment CDR3 Ig blast against germline VDJ database CDR3 CDR3

38 Summary • Recovery of CDR3 sequences of TCR a and TCR b from single T cells successful • Methods development ongoing 38

THE POWER of HUMAN™






































