Exhibit 99.2

Translating biological complexity into more powerful therapeutics January 2017

Forward Looking Statements This presentation contains forward - looking statements that are based on the company’s current expectations, assumptions, estimates and projections about the company and the pharmaceutical industry . The company makes no representations about the accuracy of such statements estimates or projections . Forward - looking statements are indicated by words such as : may, will, should, predict, continue, plan, expect, anticipate, estimate, intend, believe, could, goal objectives and similar expressions . Forward - looking statements may include, but are not limited to, statements concerning the company’s anticipated performance, including revenue and profit expectations ; development and implementation of our collaborations ; duration ; size ; scope and revenue associated with collaboration partnerships ; benefits provided to collaboration partners by our technology ; business mix ; revenues and growth in our partner base ; market opportunities ; competing technologies, industry conditions and trends ; and regulatory developments . Actual results may differ materially from the anticipated results due to substantial risks and uncertainties related to the company and the biopharmaceutical industry in which the company operates . 2

Proprietary single cell ‘ microengraving ’ technology reveals novel and valuable disease insights 3 Chris Love Associate Professor of Chemical Engineering at the Koch Institute for Integrative Cancer Research at MIT

Proprietary, functionalized microwell chip 4
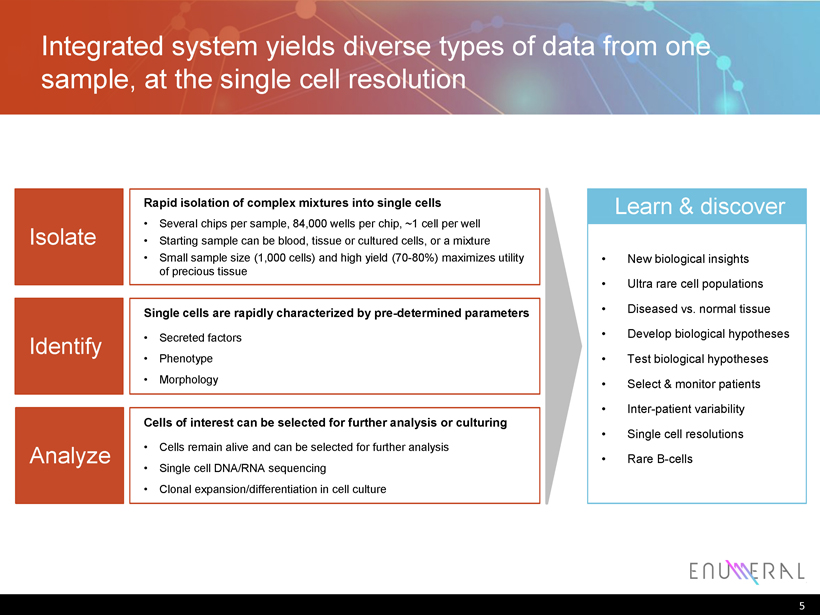
Integrated system yields diverse types of data from one sample, at the single cell resolution 5 Isolate Rapid isolation of complex mixtures into single cells • Several chips per sample, 84,000 wells per chip, ~1 cell per well • Starting sample can be blood, tissue or cultured cells, or a mixture • Small sample size (1,000 cells) and high yield (70 - 80%) maximizes utility of precious tissue Identify Single cells are rapidly characterized by pre - determined parameters • Secreted factors • Phenotype • Morphology Analyze Cells of interest can be selected for further analysis or culturing • Cells remain alive and can be selected for further analysis • Single cell DNA/RNA sequencing • Clonal expansion/differentiation in cell culture Learn & discover • New biological insights • Ultra rare cell populations • Diseased vs. normal tissue • Develop biological hypotheses • Test biological hypotheses • Select & monitor patients • Inter - patient variability • Single cell resolutions • Rare B - cells
Understanding disease at the single cell level facilitates precision medicine 6 Single cell analysis reveals new disease insights New disease insights translate into better therapeutics

ENUM past & present 2017: Unlocking full technology potential 7 Exclusive license from MIT Differentiated, high affinity PD1 antibodies discovered Tim3 antibodies discovered Focused on securing partnerships to discover & develop novel therapeutics Technology partnership SBIR Phase II contract Technology installed at MSKCC as part of NCI SBIR Commercial adaptation & industrialization (2011 - ) Internal antibody discovery (2012 - ) Unlocking full potential (2017 - ) Non - Exclusive License SBIR Phase I contract

Platform technology is automated and validated 8 Industrialized, automated & validated (2011 - 16)
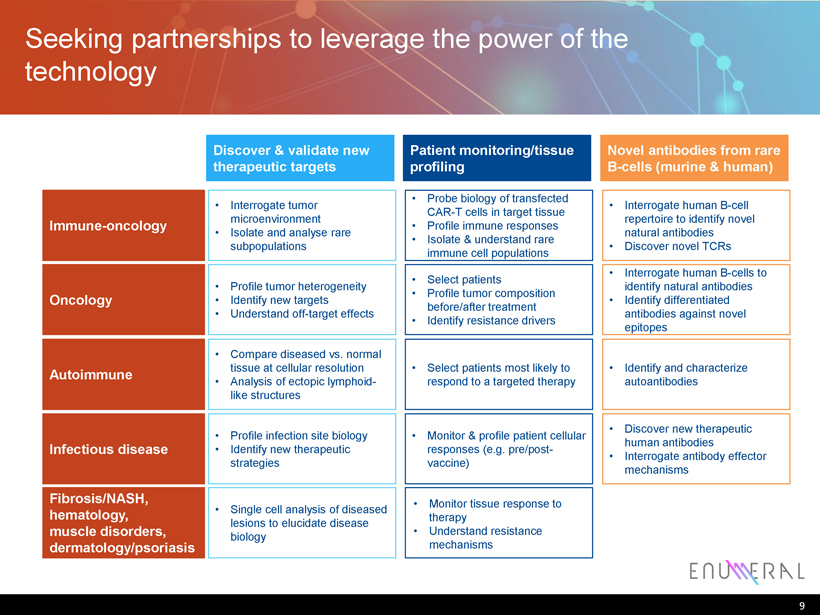
Seeking partnerships to leverage the power of the technology 9 Autoimmune • Compare diseased vs. normal tissue at cellular resolution • Analysis of ectopic lymphoid - like structures Discover & validate new therapeutic targets Immune - oncology • Interrogate tumor microenvironment • Isolate and analyse rare subpopulations Infectious disease • Profile infection site biology • Identify new therapeutic strategies Oncology • Profile tumor heterogeneity • Identify new targets • Understand off - target effects Fibrosis/NASH, hematology , muscle disorders, dermatology/psoriasis • Single cell analysis of diseased lesions to elucidate disease biology • Select patients most likely to respond to a targeted therapy Patient monitoring/tissue profiling • Probe biology of transfected CAR - T cells in target tissue • Profile immune responses • Isolate & understand rare immune cell populations • Monitor & profile patient cellular responses (e.g. pre/post - vaccine) • Select patients • Profile tumor composition before/after treatment • Identify resistance drivers • Monitor tissue response to therapy • Understand resistance mechanisms Novel antibodies from rare B - cells (murine & human) • Interrogate human B - cell repertoire to identify novel natural antibodies • Discover novel TCRs • Discover new therapeutic human antibodies • Interrogate antibody effector mechanisms • Interrogate human B - cells to identify natural antibodies • Identify differentiated antibodies against novel epitopes • Identify and characterize autoantibodies

Questions we’re being asked 10 “Can you help us understand why patients don’t respond to our drug?” “How do tumors develop resistance to our therapy?” “Can you isolate the B cells that are making the antibodies that are protecting the patient from disease?” “I’m interested in tumor - associated macrophages and tumor infiltrating lymphocytes. Can you help us isolate them?” “How does Crohn’s disease tissue differ from normal tissue?” “Can you help us understand the cellular microenvironment in fibrosis?” “What percentage of cells in a tumor are expressing our target?” “Can you help us find new i/o targets in the tumor microenvironment?”

Experienced, commercially pragmatic leadership team 11 Wael Fayad Chairman, President and CEO Robert Schaub, Ph.D. Interim Head of Research and Development Kevin G. Sarney Vice President of Finance, Treasurer, and Chief Accounting Officer Matthew A. Ebert General Counsel

Why partner with Enumeral? • Validated and versatile technology • Technology enables discovery of next generation therapeutics • Partnerships are cornerstone of company strategy • Experienced management (partnerships and science) • Flexible deal structure and terms • Rapid, streamlined deal - making with clear deliverables 12

Case studies Translating biological complexity into more powerful therapeutics

14 Discovery of new targets in the tumor microenvironment Recent clinical trials showed that targeting of inhibitory receptors on T cells induces durable responses in a subset of cancer patients, despite advanced disease. However, the regulatory switches controlling T cell function in immunosuppressive tumors are not well understood. Here we show that such inhibitory mechanisms can be systematically discovered in the tumor microenvironment. We devised an in vivo pooled shRNA screen in which shRNAs targeting negative regulators became highly enriched in tumors by releasing a block on T cell proliferation upon tumor antigen recognition. Such shRANs were identified by deep sequencing of the shRNA cassette from T cells infiltrating tumor or control tissues. One of the target genes was Ppp2r2d, a regulatory subunit of the PP2A phosphatase family: In tumors, Ppp2r2d knockdown inhibited T cell apoptosis and enhanced T cell proliferation as well as cytokine production. Key regulators of immune function can thus be discovered in relevant tissue microenvironments. Abstract • Use pooled short hairpin RNA (shRNA) libraries as discovery tools to systematically identify key modulators of T - cell proliferation in the TME in vivo • Design : Introduce shRNA pool into T - cells, activate T - cells for 48h, and identify small subset of shRNAs that restore T cell proliferation, which results in T cell accumulation within the tumor Approach • Therapeutic targets for modulating immune responses are typically identified in vitro and tested in animal models • Complex interactions of immune cells within tissues - many of which do not occur in vitro - offer untapped opportunities for therapeutic intervention Problem • How can targets for immune modulation be systematically discovered in vivo? Key Question • Establish the feasibility of in vivo discovery of novel targets for immunotherapy in complex tissue microenvironments Significance Love, JC. et al. Nature . 2014; 506(7486): 52 – 57.

15 Technology enables B - cell selection without hybridomas , enabling broader and more diverse antibody libraries Monoclonal antibodies that recognize specific antigens of interest are used as therapeutic agents and as tools for biomedical research. Discovering a single monoclonal antibody requires retrieval of an individual hybridoma from polyclonal mixtures of cells producing antibodies with a variety of specificities. The time required to isolate hybridomas by a limiting serial - dilution, however, has restricted the diversity and breadth of available antibodies. Here we present a soft lithographic method based on intaglio printing to generate microarrays comprising the secreted products of single cells. These engraved arrays enable a rapid (<12 h) and high throughput (4100,000 individual cells) system for identification, recovery and clonal expansion of cells producing antigen - specific antibodies. This method can be adapted, in principle, to detect any secreted product in a multiplexed manner. Abstract • Method involves a soft lithographic technique for microengraving that uses a dense array of microwells (0.1 – 1 nl each) containing individual cells to print a corresponding array of molecules secreted by each cell • The cells remain in culture after engraving, and the microarrays are interrogated in a manner similar to commercial microarrays of proteins or antibodies • This method enables rapid identification of those cells exhibiting desired properties, such as secretion of an antigen - specific antibody, and their subsequent recovery for clonal expansion New high throughput approach for identification, recovery & clonal expansion • Selection of hybridomas involves screening antibodies produced by large numbers of cells and retrieving those cells that produce antibodies of desired specificity. • The process is limited by requiring sufficient concentrations of antibodies for detection (may require multiple dilutions and days in culture), and the total number of manipulations limits the number of clones that can be screened efficiently in any single round of selection • Methods for the analysis of individual cells in large numbers — microfluidic devices, cell - based microarrays, enzyme - linked immunospot (ELISPOT) and hemolytic plaque assays — do not allow both high - throughput analysis of a secreted product and recovery of living cells for clonal expansion Current methods for antibody screening are time consuming and less efficient Love, JC. et al. Nature Biotechnology . 2006; 24(6):703 - 707.

16 Retrieval, expansion and characterization of single human CD8 T - cells, based on their ability to lyse a target cell CD8+ T cells are a key component of the adaptive immune response to viral infection. An inadequate CD8+ T cell response is thought to be partly responsible for the persistent chronic infection that arises following infection with HIV. It is therefore critical to identify ways to define what constitutes an adequate or inadequate response. IFN - γ production has been used as a measure of T cell function, but the relationship between cytokine production and the ability of a cell to lyse virus - infected cells is not clear. Moreover, the ability to assess multiple CD8+ T cell functions with single - cell resolution using freshly isolated blood samples, and subsequently to recover these cells for further functional analyses, has not been achieved. As described here, to address this need, we have developed a high - throughput, automated assay in 125 - pl microwells to simultaneously evaluate the ability of thousands of individual CD8+ T cells from HIV - infected patients to mediate lysis and to produce cytokines. This concurrent, direct analysis enabled us to investigate the correlation between immediate cytotoxic activity and short - term cytokine secretion. The majority of in vivo primed, circulating HIV - specific CD8+ T cells were discordant for cytolysis and cytokine secretion, notably IFN - γ, when encountering cognate antigen presented on defined numbers of cells. Our approach should facilitate determination of signatures of functional variance among individual effector CD8+ T cells, including those from mucosal samples and those induced by vaccines. Abstract • Antigen - specific CD8+ T cells can exhibit a range of functions, including the production of effector cytokines, degranulation, cytolytic activity, suppression of viral replication, and proliferation upon exposure to cells presenting antigen • The importance of CD8+ T cells for controlling replication in chronic human viral infections has motivated extensive research to define phenotypic and functional attributes of an effective response Problem • Identify ways to define what constitutes an adequate or inadequate CD8+ T cell response. Key Question • High - throughput single - cell assay to directly observe cytolytic activity and cytokine secretion ex vivo. a new tool for evaluating multiple functional activities associated with antigen - specific primary T cells following encounter with one or a small number of APCs Significance • High throughput microwell single cell assay for cytolysis to understand the direct relationship between cytolysis and other functional responses by T cells, while preserving the ability to then further characterize specific reactive cells Approach Varadarajan , N. et al. Journal of Clinical Investigation . 2011; 121(11):4322 - 4331.

17 Understanding the temporal dynamics of T - cell activation at the single cell level The release of cytokines by T cells defines a significant part of their functional activity in vivo, and their ability to produce multiple cytokines has been associated with beneficial immune responses. To date, time - integrated end - point measurements have obscured whether these polyfunctional states arise from the simultaneous or successive release of cytokines. Here, we used serial, time - dependent, single - cell analysis of primary human T cells to resolve the temporal dynamics of cytokine secretion from individual cells after activation ex vivo. We show that multifunctional, Th1 - skewed cytokine responses (IFN - γ, IL - 2, TNFα) are initiated asynchronously, but the ensuing dynamic trajectories of these responses evolve programmatically in a sequential manner. That is, cells predominantly release one of these cytokines at a time rather than maintain active secretion of multiple cytokines simultaneously. Furthermore, these dynamic trajectories are strongly associated with the various states of cell differentiation suggesting that transient programmatic activities of many individual T cells contribute to sustained, population - level responses. The trajectories of responses by single cells may also provide unique, time - dependent signatures for immune monitoring that are less compromised by the timing and duration of integrated measures. Abstract • Examine how the synchrony and evolution of secreted cytokines varies upon activation among different subsets of primary human CD3+ T cells isolated from peripheral blood. • Design : Using a combination of imaging cytometry and quantitative single - cell analysis of secreted cytokines, we monitored the release of three Th1 - skewed cytokines (IFN - γ, IL - 2, and TNFα) over time Approach • The production of multiple cytokines by T cells has been associated with productive immune responses to infectious diseases (5 – 7) and to vaccines • The manner in which polyfunctional responses by individual cells contribute to the evolution of an immune response at a population level is not well understood Problem • Aim to clarify when activated T cells initiate the release of cytokines, and how their responses evolve in time Key Objective • Found that T cells initiate the release of cytokines at different points in time upon stimulation and that most of these cells initiate secretion in a monofunctional state • Dynamic monitoring of immune cells may improve profiling functional responses associated with immune status relative to integrated, endpoint measurements Significance Han, Q. et al. PNAS . 2012; 109(5):1607 - 1612.

Single cell profiling of diseased Crohn’s vs. normal tissue elucidated new therapeutic strategies 18 99% 1% Normal tissue, adjacent to diseased tissue: low expression Non-Secretors Secretors 0 20 40 60 80 100 120 IFN-g IL17 IFN-g/IL17 Cell Number Cytokine Active Crohn’s disease tissue Non-Secretors Secretors 0 20 40 60 80 100 120 IFN-g IL17 IFN-g/IL17 Cell Number Cytokine
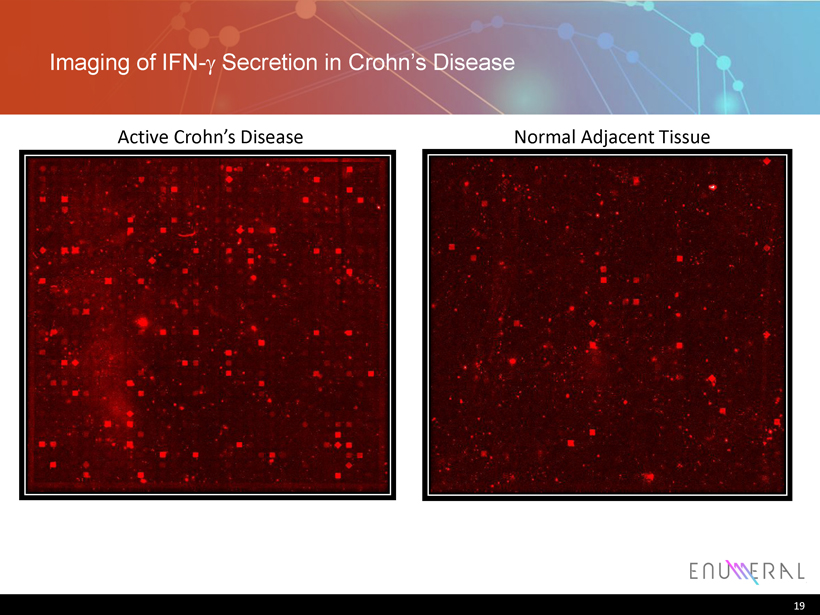
Imaging of IFN - g Secretion in Crohn’s Disease 19 Active Crohn’s Disease Normal Adjacent Tissue
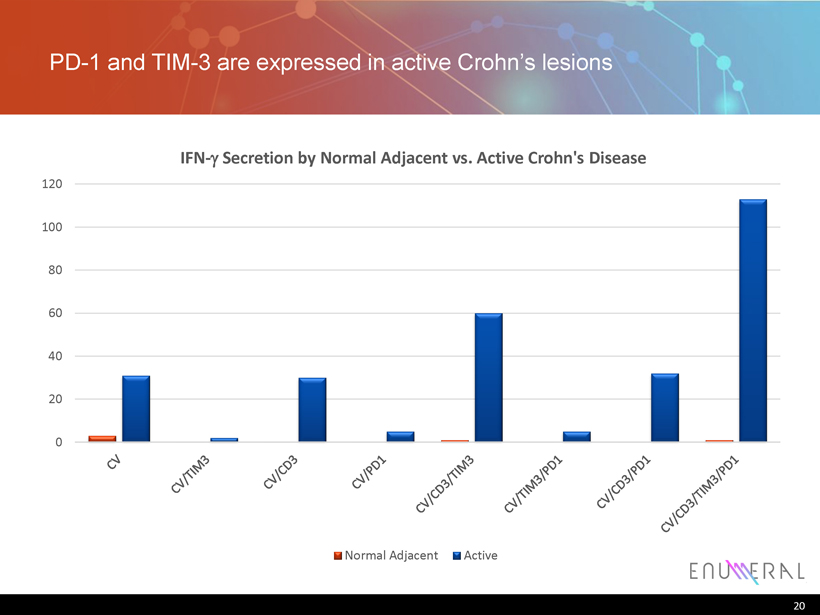
PD - 1 and TIM - 3 are expressed in active Crohn’s lesions 20 0 20 40 60 80 100 120 IFN - g Secretion by Normal Adjacent vs. Active Crohn's Disease Normal Adjacent Active
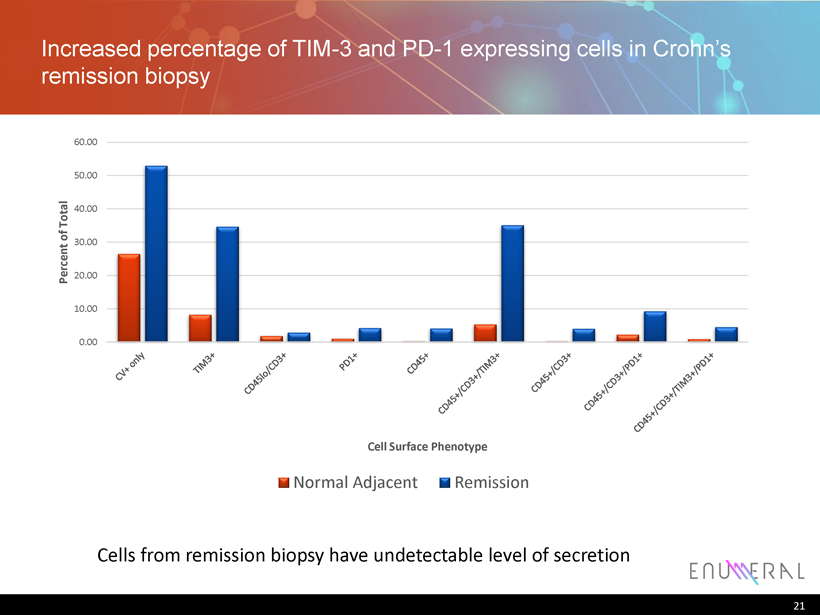
Increased percentage of TIM - 3 and PD - 1 expressing cells in Crohn’s remission biopsy 21 0.00 10.00 20.00 30.00 40.00 50.00 60.00 Percent of Total Cell Surface Phenotype Normal Adjacent Remission Cells from remission biopsy have undetectable level of secretion
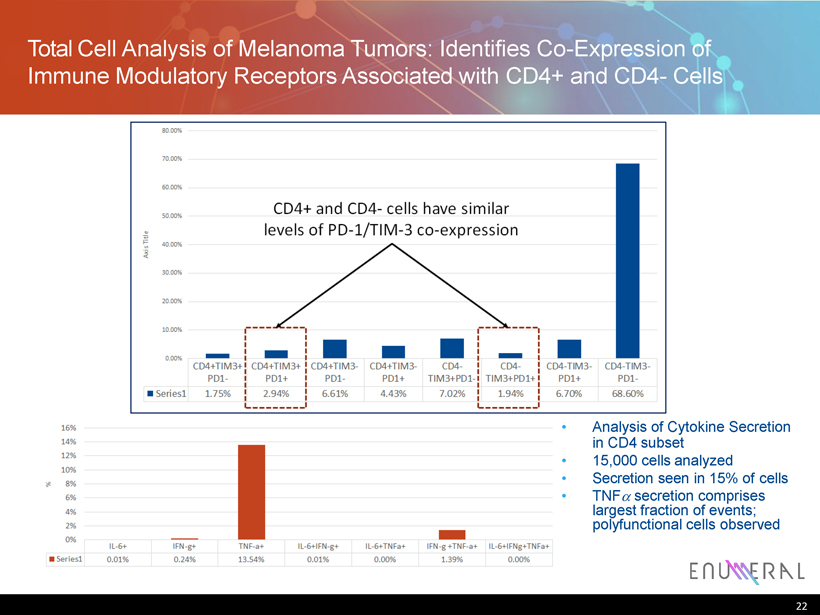
Total C ell A nalysis of Melanoma Tumors : Identifies C o - E xpression of Immune Modulatory Receptors Associated with CD4+ and CD4 - Cells • Analysis of Cytokine Secretion in CD4 subset • 15,000 cells analyzed • Secretion seen in 15% of cells • TNF secretion comprises largest fraction of events; polyfunctional cells observed 22
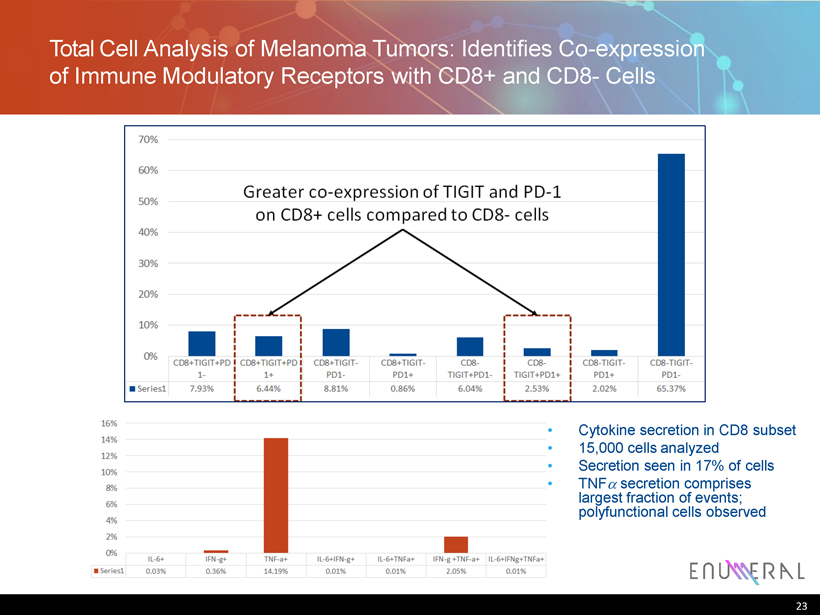
Total C ell A nalysis of Melanoma Tumors : Identifies C o - expression of Immune Modulatory Receptors with CD8 + and CD8 - Cells • Cytokine secretion in CD8 subset • 15,000 cells analyzed • Secretion seen in 17% of cells • TNF secretion comprises largest fraction of events; polyfunctional cells observed 23

In - house antibody discovery program Discovering broad and diverse antibodies against targets of high interest
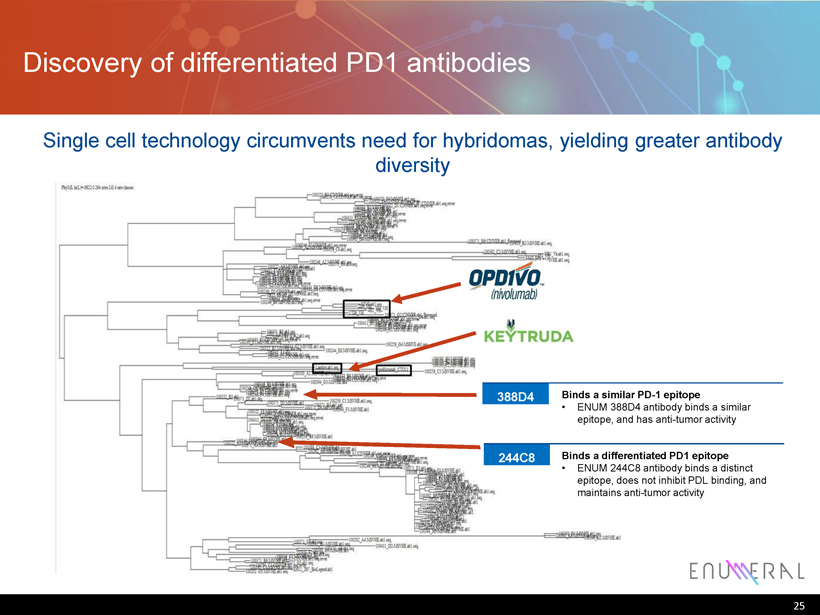
Discovery of differentiated PD1 antibodies 25 244C8 Binds a differentiated PD1 epitope • ENUM 244C8 antibody binds a distinct epitope, does not inhibit PDL binding, and maintains anti - tumor activity 388D4 Binds a similar PD - 1 epitope • ENUM 388D4 antibody binds a similar epitope, and has anti - tumor activity Single cell technology circumvents need for hybridomas , yielding greater antibody diversity
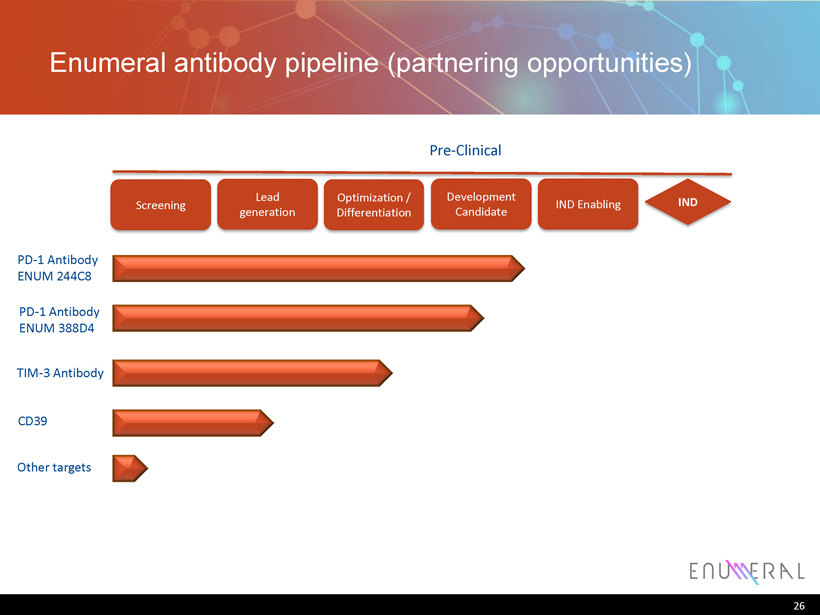
Enumeral antibody pipeline (partnering opportunities) 26 PD - 1 Antibody ENUM 244C8 PD - 1 Antibody ENUM 388D4 TIM - 3 Antibody CD39 Lead generation Screening Optimization / Differentiation Pre - Clinical IND Enabling IND Other targets Development Candidate

























