
Analyst & Investor Research Webcast August 25, 2020 Exhibit 99.1

Forward-looking statements This document contains forward-looking statements. All statements other than statements of historical facts contained in this document, including statements regarding possible or assumed future results of operations, preclinical and clinical studies, business strategies, research and development plans, collaborations and partnerships, regulatory activities and timing thereof, competitive position, potential growth opportunities, use of proceeds and the effects of competition are forward-looking statements. These statements involve known and unknown risks, uncertainties and other important factors that may cause the actual results, performance or achievements of Wave Life Sciences Ltd. (the “Company”) to be materially different from any future results, performance or achievements expressed or implied by the forward-looking statements. In some cases, you can identify forward-looking statements by terms such as “may,” “will,” “should,” “expect,” “plan,” “aim,” “anticipate,” “could,” “intend,” “target,” “project,” “contemplate,” “believe,” “estimate,” “predict,” “potential” or “continue” or the negative of these terms or other similar expressions. The forward-looking statements in this presentation are only predictions. The Company has based these forward-looking statements largely on its current expectations and projections about future events and financial trends that it believes may affect the Company’s business, financial condition and results of operations. These forward-looking statements speak only as of the date of this presentation and are subject to a number of risks, uncertainties and assumptions, including those listed under Risk Factors in the Company’s Form 10-K and other filings with the SEC, some of which cannot be predicted or quantified and some of which are beyond the Company’s control. The events and circumstances reflected in the Company’s forward-looking statements may not be achieved or occur, and actual results could differ materially from those projected in the forward-looking statements. Moreover, the Company operates in a dynamic industry and economy. New risk factors and uncertainties may emerge from time to time, and it is not possible for management to predict all risk factors and uncertainties that the Company may face. Except as required by applicable law, the Company does not plan to publicly update or revise any forward-looking statements contained herein, whether as a result of any new information, future events, changed circumstances or otherwise.

Paul Bolno, MD, MBA President and CEO Vision & Strategy Chandra Vargeese, PhD Chief Technology Officer PRISM Platform Update ADAR Editing Kenneth Rhodes, PhD SVP, Therapeutics Discovery Neurology Pipeline C9orf72 Program Today’s speakers Conclusion and Q&A

Vision and Strategy Paul Bolno, MD, MBA President and CEO

Wave Life Sciences VISION We envision a future in which the diagnosis of a genetically-defined disease leads to effective and available treatment, providing patients and their families the ability to realize a brighter future MISSION Apply innovative nucleic acid chemistry and deep biological insights to develop transformative medicines for millions of people living with devastating conditions Building a fully integrated genetic medicines company
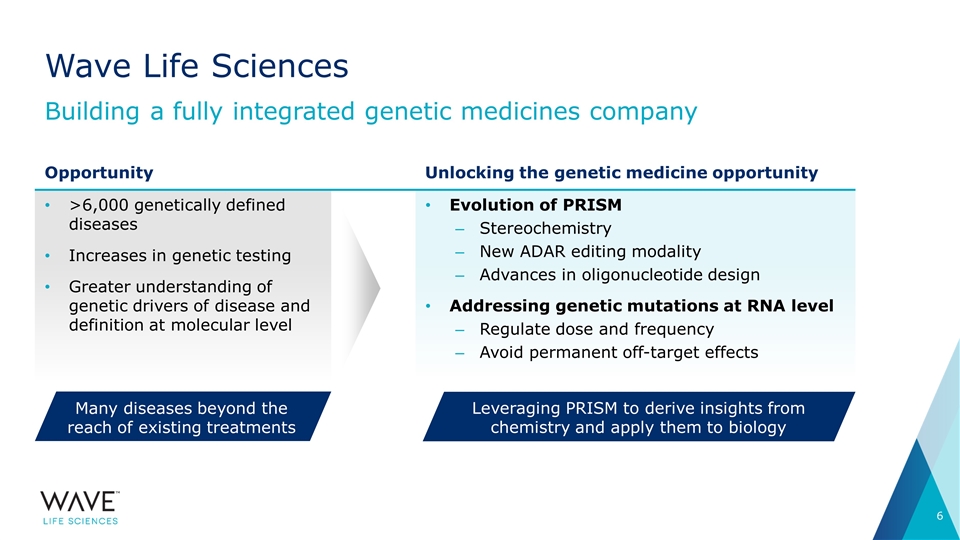
Wave Life Sciences Building a fully integrated genetic medicines company >6,000 genetically defined diseases Increases in genetic testing Greater understanding of genetic drivers of disease and definition at molecular level Evolution of PRISM Stereochemistry New ADAR editing modality Advances in oligonucleotide design Addressing genetic mutations at RNA level Regulate dose and frequency Avoid permanent off-target effects Opportunity Unlocking the genetic medicine opportunity Leveraging PRISM to derive insights from chemistry and apply them to biology Many diseases beyond the reach of existing treatments

PRISM platform designed to achieve four goals ü ü ü ü Enable multiple modalities Silencing, Splicing, ADAR editing Ability to optimize pharmacology Potency, Exposure, Durability Target engagement across key tissues Scalable and cost-effective manufacturing
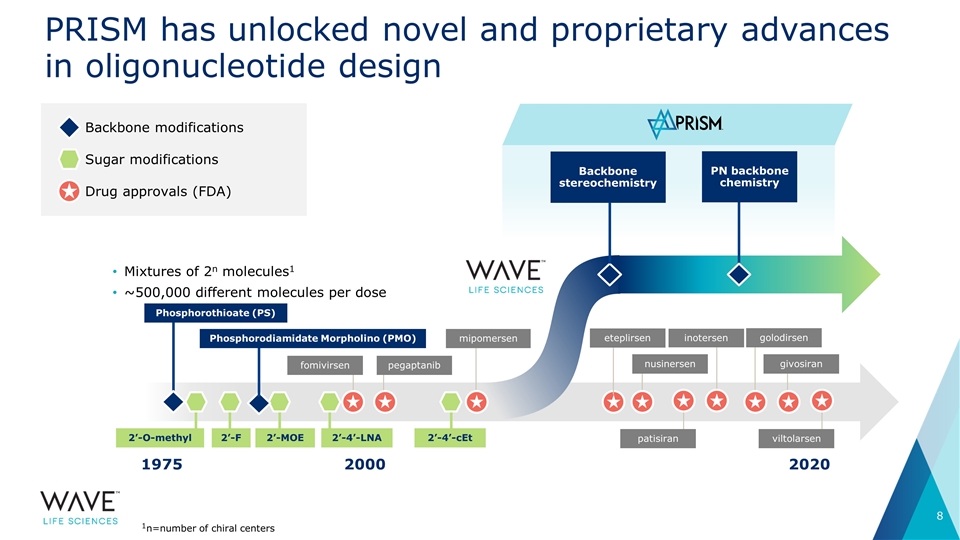
PRISM has unlocked novel and proprietary advances in oligonucleotide design Backbone modifications Sugar modifications Drug approvals (FDA) 1975 2020 2000 Mixtures of 2n molecules1 ~500,000 different molecules per dose fomivirsen pegaptanib Phosphorothioate (PS) mipomersen nusinersen PN backbone chemistry Backbone stereochemistry 2’-4’-cEt 2’-O-methyl 2’-F 2’-4’-LNA 1n=number of chiral centers 2’-MOE Phosphorodiamidate Morpholino (PMO) eteplirsen golodirsen givosiran patisiran inotersen viltolarsen
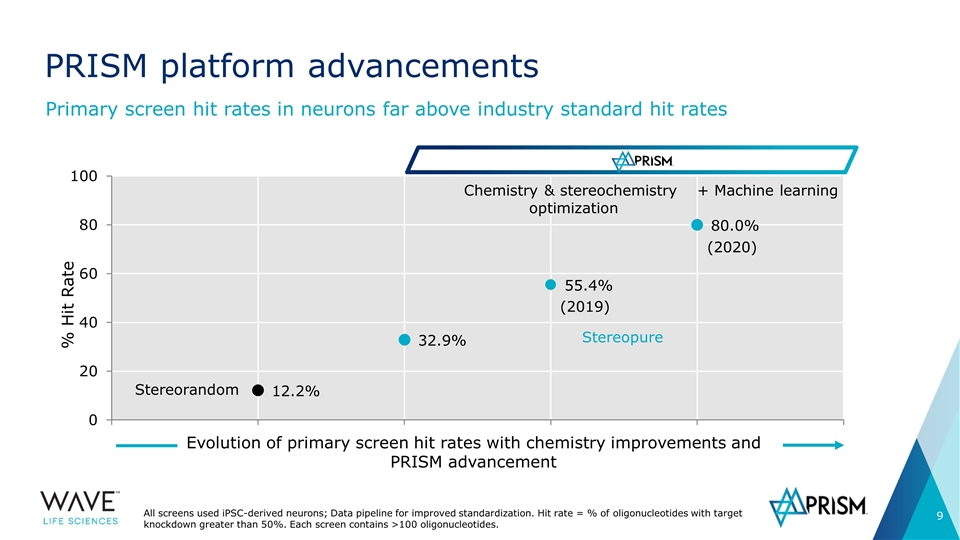
PRISM platform advancements Primary screen hit rates in neurons far above industry standard hit rates Stereorandom Chemistry & stereochemistry optimization + Machine learning Stereopure Evolution of primary screen hit rates with chemistry improvements and PRISM advancement All screens used iPSC-derived neurons; Data pipeline for improved standardization. Hit rate = % of oligonucleotides with target knockdown greater than 50%. Each screen contains >100 oligonucleotides. (2019) (2020)
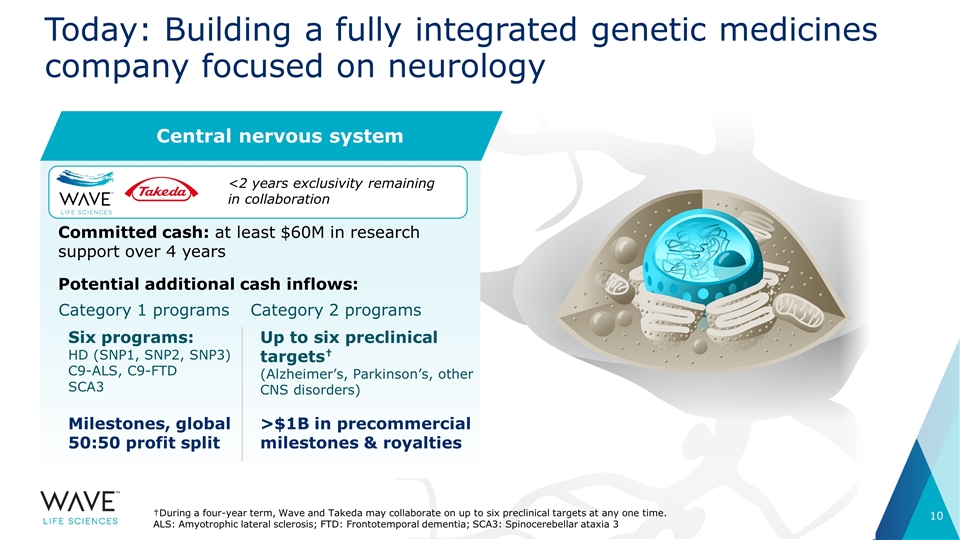
Today: Building a fully integrated genetic medicines company focused on neurology Committed cash: at least $60M in research support over 4 years Potential additional cash inflows: Category 1 programs Six programs: HD (SNP1, SNP2, SNP3) C9-ALS, C9-FTD SCA3 Milestones, global 50:50 profit split Category 2 programs Up to six preclinical targets† (Alzheimer’s, Parkinson’s, other CNS disorders) >$1B in precommercial milestones & royalties †During a four-year term, Wave and Takeda may collaborate on up to six preclinical targets at any one time. ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia; SCA3: Spinocerebellar ataxia 3 Central nervous system <2 years exclusivity remaining in collaboration
Looking ahead: Building a fully integrated genetic medicines company focused on neurology Employing new chemistries and modalities to expand wholly-owned neurology pipeline ADAR editing to access new target classes and new pathways PRISM enables access to larger set of potential indications than other existing platforms Neurology pipeline expansion
Leveraging platform discovery research to build out areas of potential new biology Hepatic diseases Ophthalmology Muscle diseases Additional therapeutic areas Opportunities outside of neurology
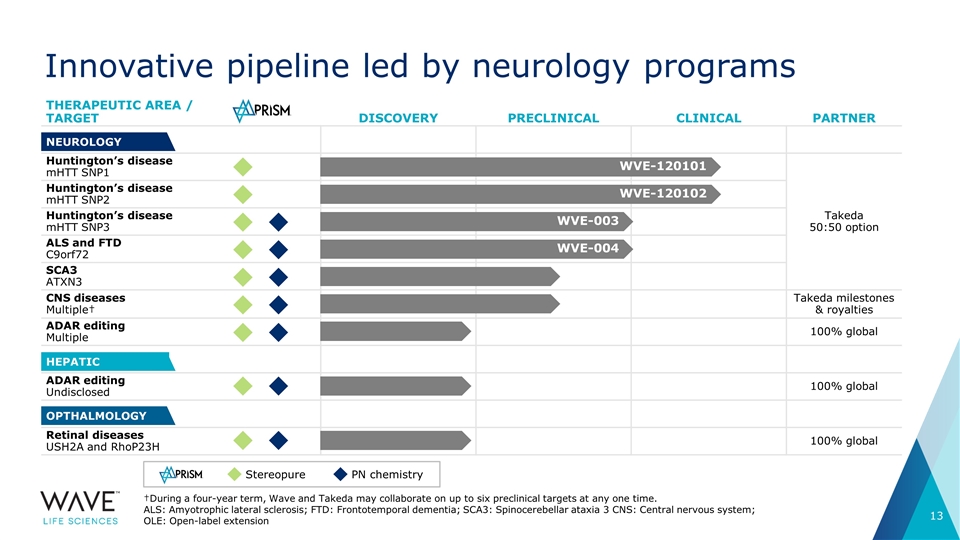
THERAPEUTIC AREA / TARGET DISCOVERY PRECLINICAL CLINICAL PARTNER Huntington’s disease mHTT SNP1 Takeda 50:50 option Huntington’s disease mHTT SNP2 Huntington’s disease mHTT SNP3 ALS and FTD C9orf72 SCA3 ATXN3 CNS diseases Multiple† Takeda milestones & royalties ADAR editing Multiple 100% global ADAR editing Undisclosed 100% global Retinal diseases USH2A and RhoP23H 100% global NEUROLOGY HEPATIC OPTHALMOLOGY WVE-003 WVE-004 WVE-120101 WVE-120102 Innovative pipeline led by neurology programs †During a four-year term, Wave and Takeda may collaborate on up to six preclinical targets at any one time. ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia; SCA3: Spinocerebellar ataxia 3 CNS: Central nervous system; OLE: Open-label extension Stereopure PN chemistry

Wave Life Sciences: Redefining the potential of RNA therapeutics in neurology Continuous learning Platform engine delivering new targets Four global clinical neurology programs expected next year, with multiple data readouts by 2022 Positioned to deliver multiple clinical trial applications over the next three years Leveraging platform to bring new neurology targets, including editing targets in CNS, to clinic Collaborations to unlock further value Well positioned to drive near-term value from PRISM

PRISM platform update Chandra Vargeese, PhD Chief Technology Officer
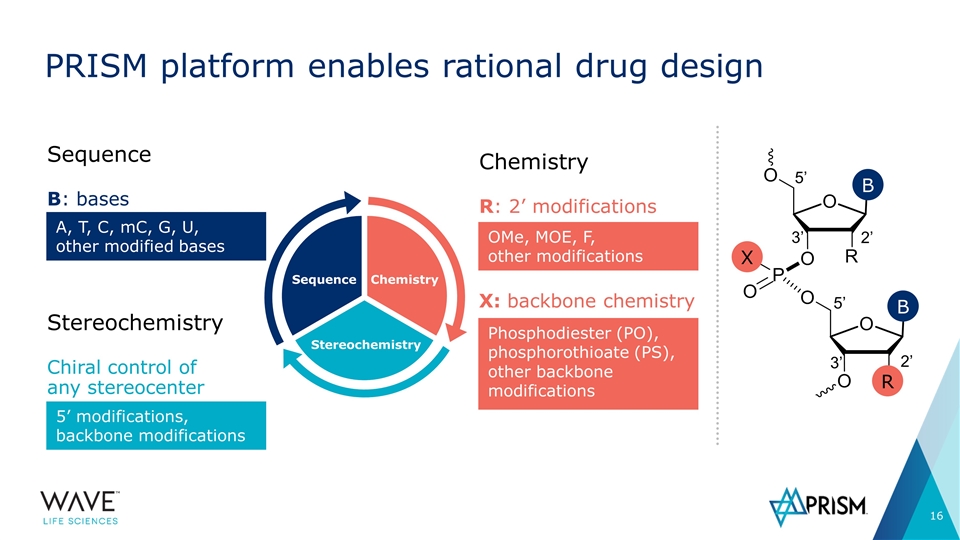
Sequence Stereochemistry Chemistry PRISM platform enables rational drug design Chemistry R: 2’ modifications OMe, MOE, F, other modifications 5’ 2’ 3’ 5’ 3’ 2’ R X B B X: backbone chemistry Phosphodiester (PO), phosphorothioate (PS), other backbone modifications Sequence B: bases A, T, C, mC, G, U, other modified bases Stereochemistry Chiral control of any stereocenter 5’ modifications, backbone modifications
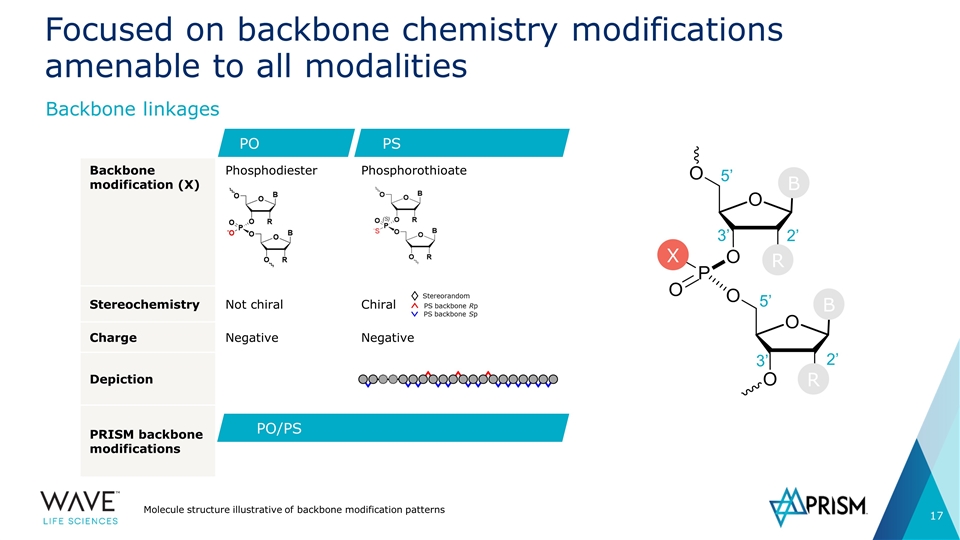
Backbone modification (X) Phosphodiester Phosphorothioate Stereochemistry Not chiral Chiral Charge Negative Negative Depiction PRISM backbone modifications Focused on backbone chemistry modifications amenable to all modalities Molecule structure illustrative of backbone modification patterns Backbone linkages PS PO PO/PS Stereorandom PS backbone Rp PS backbone Sp 5’ 2’ 3’ 5’ 3’ 2’ R X B B R
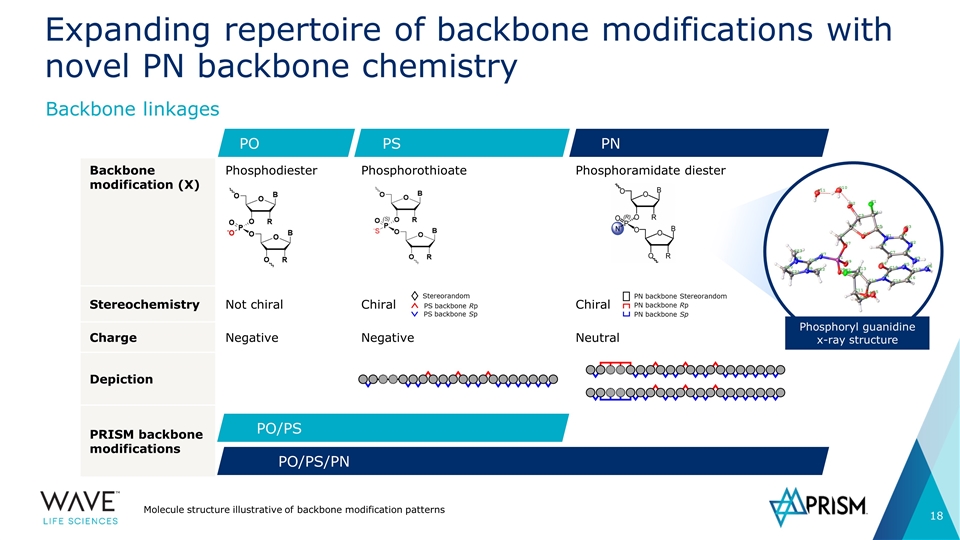
Backbone modification (X) Phosphodiester Phosphorothioate Phosphoramidate diester Stereochemistry Not chiral Chiral Chiral Charge Negative Negative Neutral Depiction PRISM backbone modifications Expanding repertoire of backbone modifications with novel PN backbone chemistry Molecule structure illustrative of backbone modification patterns Backbone linkages PS PO PN PO/PS PO/PS/PN Phosphoryl guanidine x-ray structure Stereorandom PS backbone Rp PS backbone Sp PN backbone Sp PN backbone Rp PN backbone Stereorandom
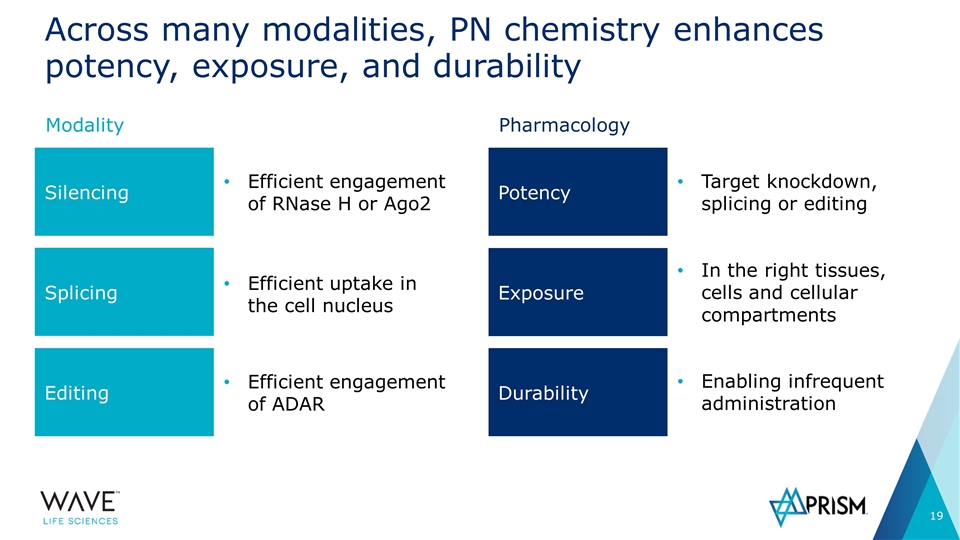
Across many modalities, PN chemistry enhances potency, exposure, and durability Modality Pharmacology Potency Exposure Durability Silencing Splicing Editing Efficient engagement of RNase H or Ago2 Efficient uptake in the cell nucleus Efficient engagement of ADAR Enabling infrequent administration In the right tissues, cells and cellular compartments Target knockdown, splicing or editing
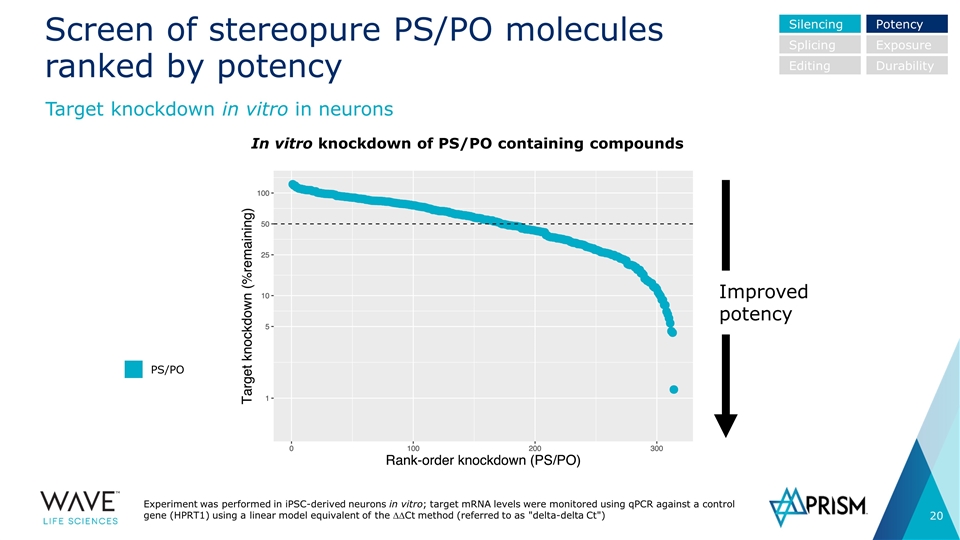
Screen of stereopure PS/PO molecules ranked by potency Experiment was performed in iPSC-derived neurons in vitro; target mRNA levels were monitored using qPCR against a control gene (HPRT1) using a linear model equivalent of the DDCt method (referred to as "delta-delta Ct") In vitro knockdown of PS/PO containing compounds Silencing Splicing Editing Potency Exposure Durability PS/PO Improved potency Target knockdown in vitro in neurons
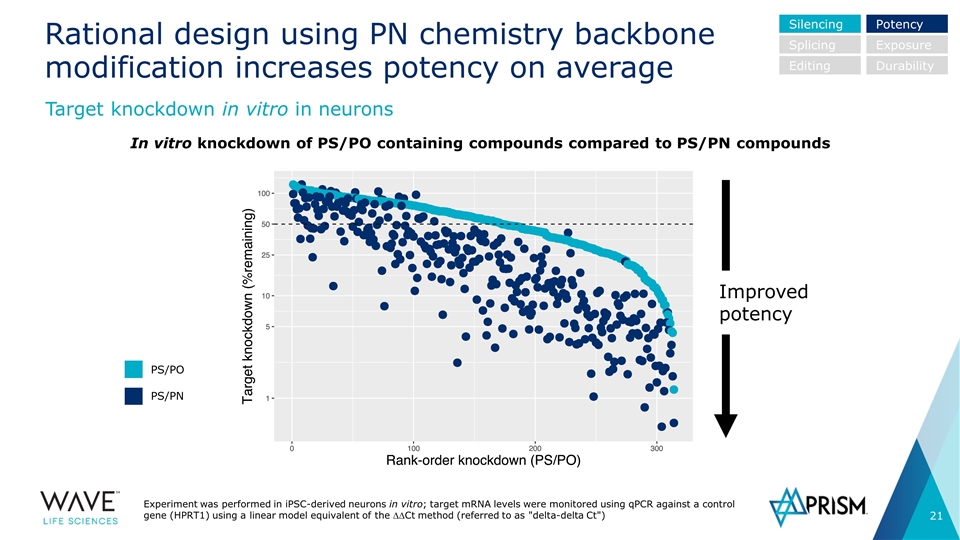
In vitro knockdown of PS/PO containing compounds compared to PS/PN compounds Rational design using PN chemistry backbone modification increases potency on average Experiment was performed in iPSC-derived neurons in vitro; target mRNA levels were monitored using qPCR against a control gene (HPRT1) using a linear model equivalent of the DDCt method (referred to as "delta-delta Ct") Target knockdown in vitro in neurons Silencing Splicing Editing Potency Exposure Durability PS/PO PS/PN Improved potency
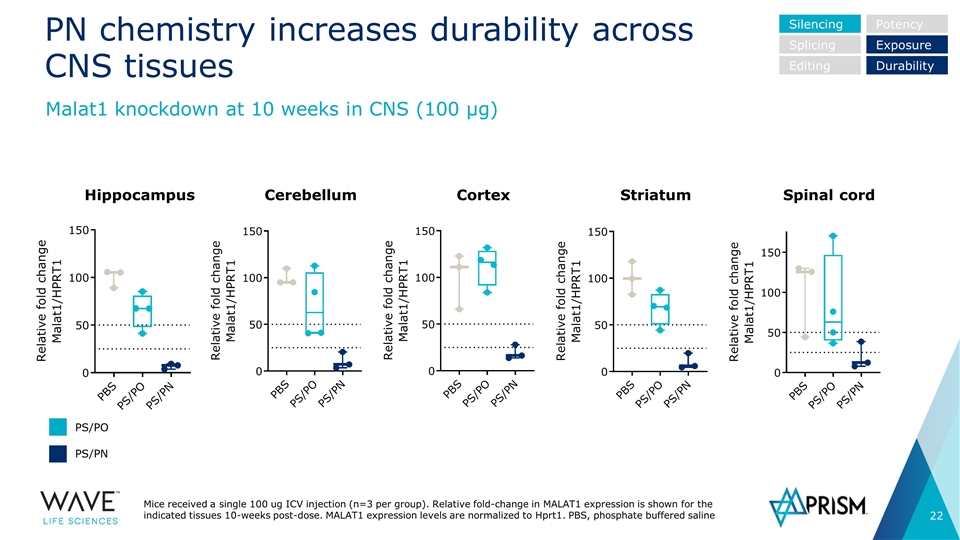
PN chemistry increases durability across CNS tissues Mice received a single 100 ug ICV injection (n=3 per group). Relative fold-change in MALAT1 expression is shown for the indicated tissues 10-weeks post-dose. MALAT1 expression levels are normalized to Hprt1. PBS, phosphate buffered saline Malat1 knockdown at 10 weeks in CNS (100 µg) Hippocampus Cerebellum Spinal cord Cortex Striatum Silencing Splicing Editing Potency Exposure Durability PS/PO PS/PN
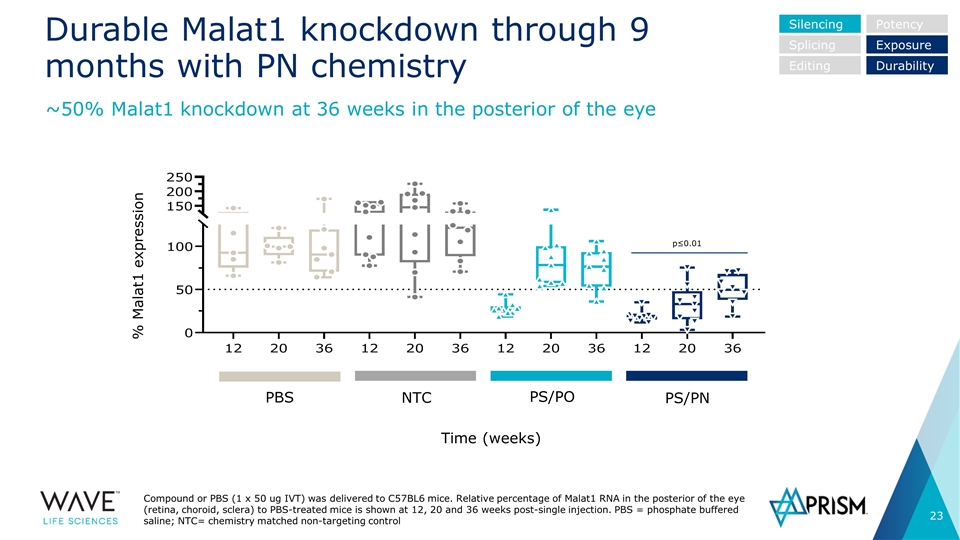
Durable Malat1 knockdown through 9 months with PN chemistry Compound or PBS (1 x 50 ug IVT) was delivered to C57BL6 mice. Relative percentage of Malat1 RNA in the posterior of the eye (retina, choroid, sclera) to PBS-treated mice is shown at 12, 20 and 36 weeks post-single injection. PBS = phosphate buffered saline; NTC= chemistry matched non-targeting control ~50% Malat1 knockdown at 36 weeks in the posterior of the eye PBS NTC PS/PO PS/PN Silencing Splicing Editing Potency Exposure Durability % Malat1 expression Time (weeks) p≤0.01
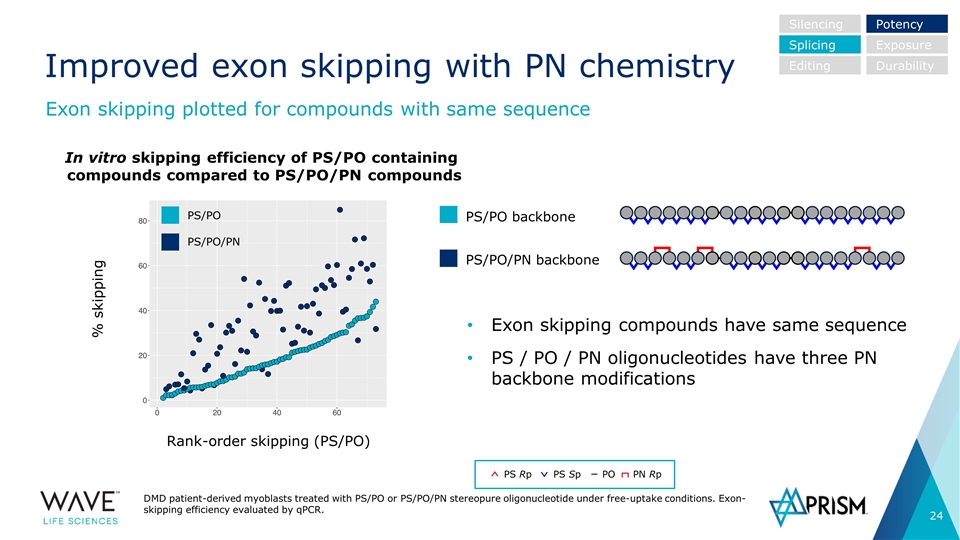
Improved exon skipping with PN chemistry DMD patient-derived myoblasts treated with PS/PO or PS/PO/PN stereopure oligonucleotide under free-uptake conditions. Exon-skipping efficiency evaluated by qPCR. Exon skipping plotted for compounds with same sequence In vitro skipping efficiency of PS/PO containing compounds compared to PS/PO/PN compounds Exon skipping compounds have same sequence PS / PO / PN oligonucleotides have three PN backbone modifications Silencing Splicing Editing Potency Exposure Durability % skipping Rank-order skipping (PS/PO) PS/PO PS/PO/PN PS/PO backbone PS/PO/PN backbone PS Rp PS Sp PO PN Rp
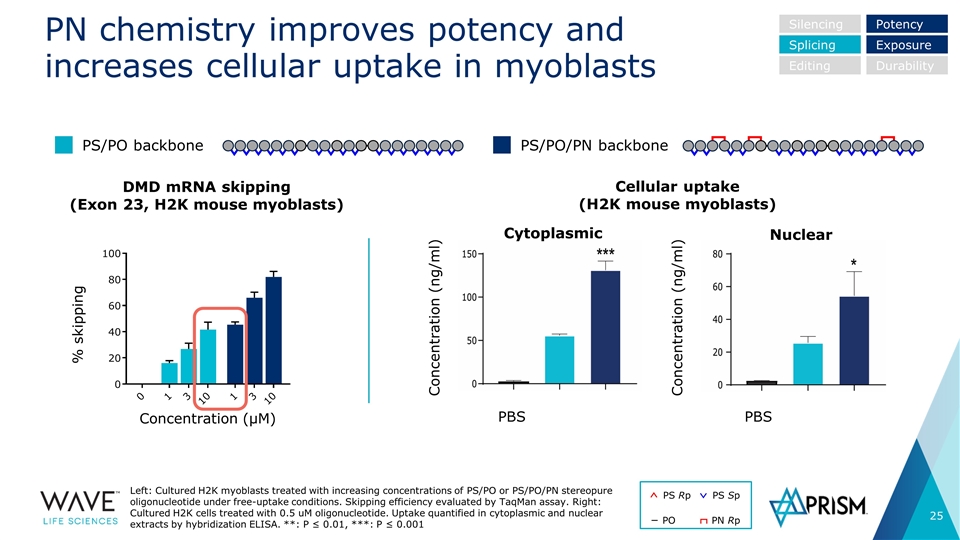
PN chemistry improves potency and increases cellular uptake in myoblasts Left: Cultured H2K myoblasts treated with increasing concentrations of PS/PO or PS/PO/PN stereopure oligonucleotide under free-uptake conditions. Skipping efficiency evaluated by TaqMan assay. Right: Cultured H2K cells treated with 0.5 uM oligonucleotide. Uptake quantified in cytoplasmic and nuclear extracts by hybridization ELISA. **: P ≤ 0.01, ***: P ≤ 0.001 DMD mRNA skipping (Exon 23, H2K mouse myoblasts) % skipping Concentration (µM) Concentration (ng/ml) Concentration (ng/ml) PBS PBS Cellular uptake (H2K mouse myoblasts) PS/PO backbone PS/PO/PN backbone Cytoplasmic Nuclear Silencing Splicing Editing Potency Exposure Durability PS Rp PS Sp PO PN Rp
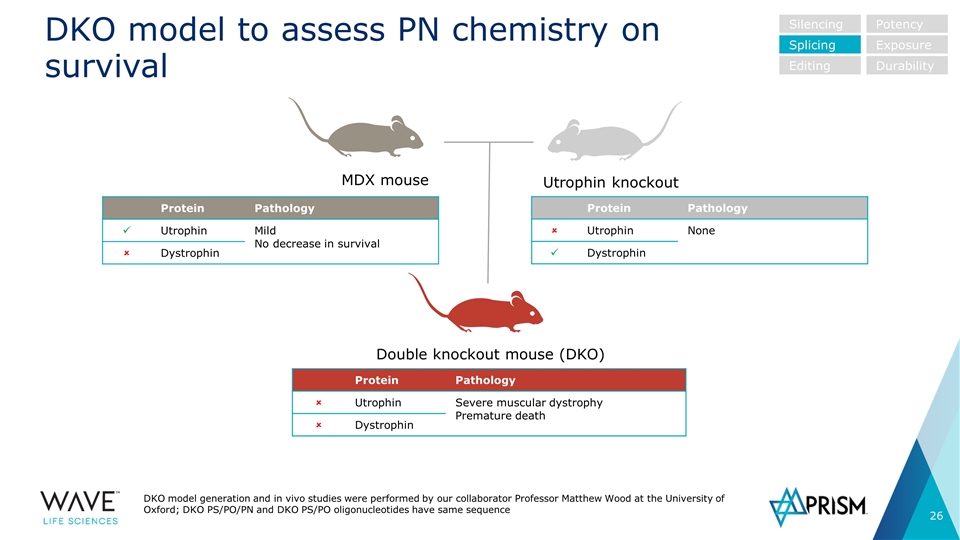
DKO model to assess PN chemistry on survival DKO model generation and in vivo studies were performed by our collaborator Professor Matthew Wood at the University of Oxford; DKO PS/PO/PN and DKO PS/PO oligonucleotides have same sequence Double knockout mouse (DKO) Protein Pathology ü Utrophin Mild No decrease in survival û Dystrophin Protein Pathology û Utrophin None ü Dystrophin MDX mouse Protein Pathology û Utrophin Severe muscular dystrophy Premature death û Dystrophin Utrophin knockout Silencing Splicing Editing Potency Exposure Durability
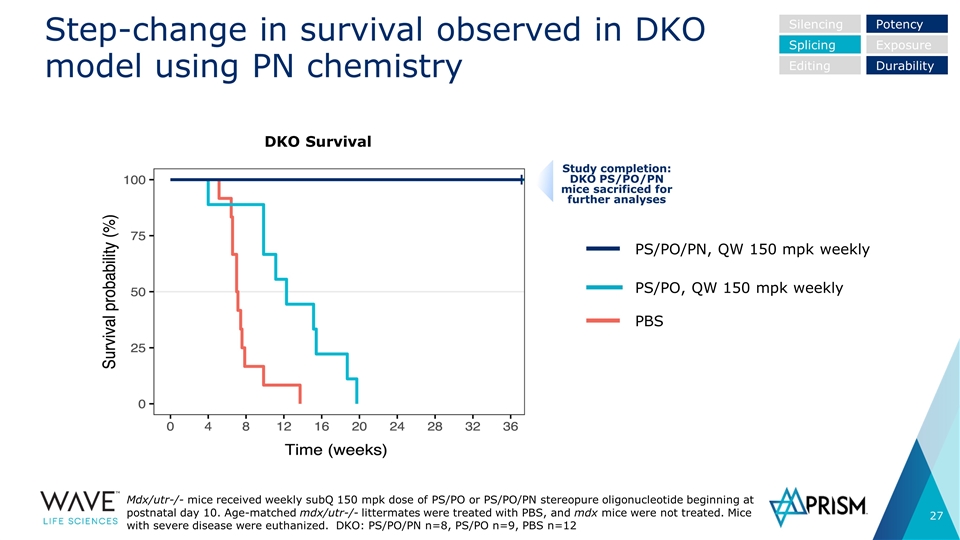
Study completion: DKO PS/PO/PN mice sacrificed for further analyses Step-change in survival observed in DKO model using PN chemistry Mdx/utr-/- mice received weekly subQ 150 mpk dose of PS/PO or PS/PO/PN stereopure oligonucleotide beginning at postnatal day 10. Age-matched mdx/utr-/- littermates were treated with PBS, and mdx mice were not treated. Mice with severe disease were euthanized. DKO: PS/PO/PN n=8, PS/PO n=9, PBS n=12 PBS PS/PO, QW 150 mpk weekly PS/PO/PN, QW 150 mpk weekly Silencing Splicing Editing Potency Exposure Durability DKO Survival
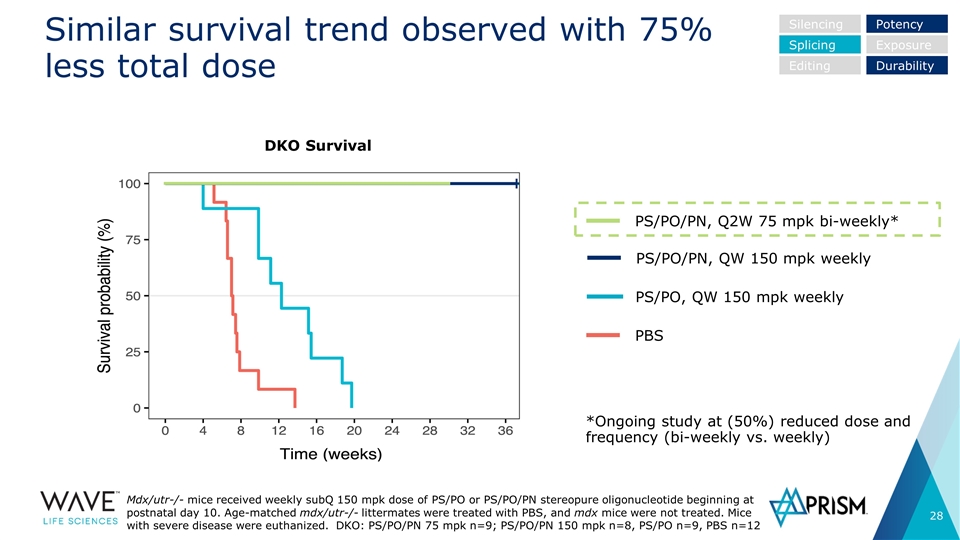
Similar survival trend observed with 75% less total dose PBS PS/PO, QW 150 mpk weekly PS/PO/PN, QW 150 mpk weekly PS/PO/PN, Q2W 75 mpk bi-weekly* Silencing Splicing Editing Potency Exposure Durability *Ongoing study at (50%) reduced dose and frequency (bi-weekly vs. weekly) DKO Survival Mdx/utr-/- mice received weekly subQ 150 mpk dose of PS/PO or PS/PO/PN stereopure oligonucleotide beginning at postnatal day 10. Age-matched mdx/utr-/- littermates were treated with PBS, and mdx mice were not treated. Mice with severe disease were euthanized. DKO: PS/PO/PN 75 mpk n=9; PS/PO/PN 150 mpk n=8, PS/PO n=9, PBS n=12
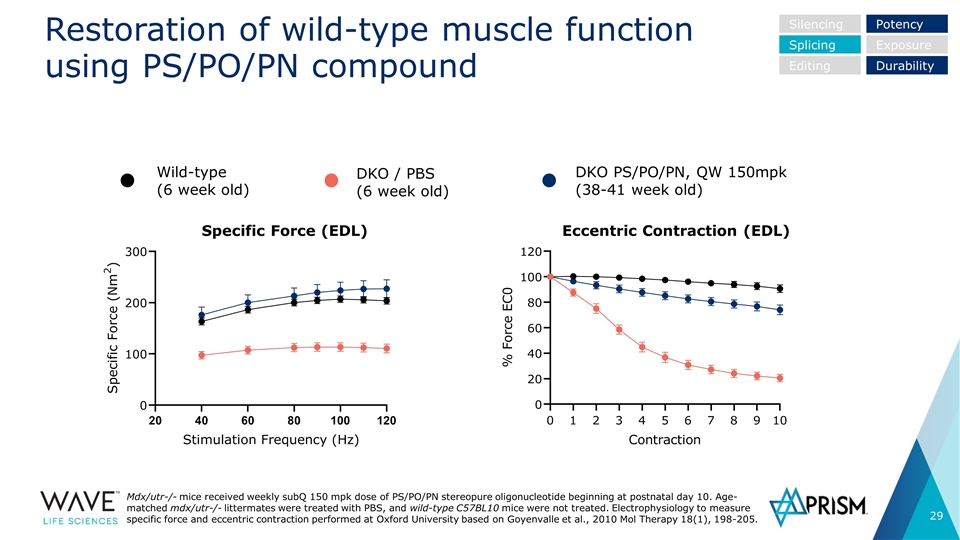
Restoration of wild-type muscle function using PS/PO/PN compound DKO / PBS (6 week old) DKO PS/PO/PN, QW 150mpk (38-41 week old) Wild-type (6 week old) Specific Force (EDL) Eccentric Contraction (EDL) Silencing Splicing Editing Potency Exposure Durability Mdx/utr-/- mice received weekly subQ 150 mpk dose of PS/PO/PN stereopure oligonucleotide beginning at postnatal day 10. Age-matched mdx/utr-/- littermates were treated with PBS, and wild-type C57BL10 mice were not treated. Electrophysiology to measure specific force and eccentric contraction performed at Oxford University based on Goyenvalle et al., 2010 Mol Therapy 18(1), 198-205.
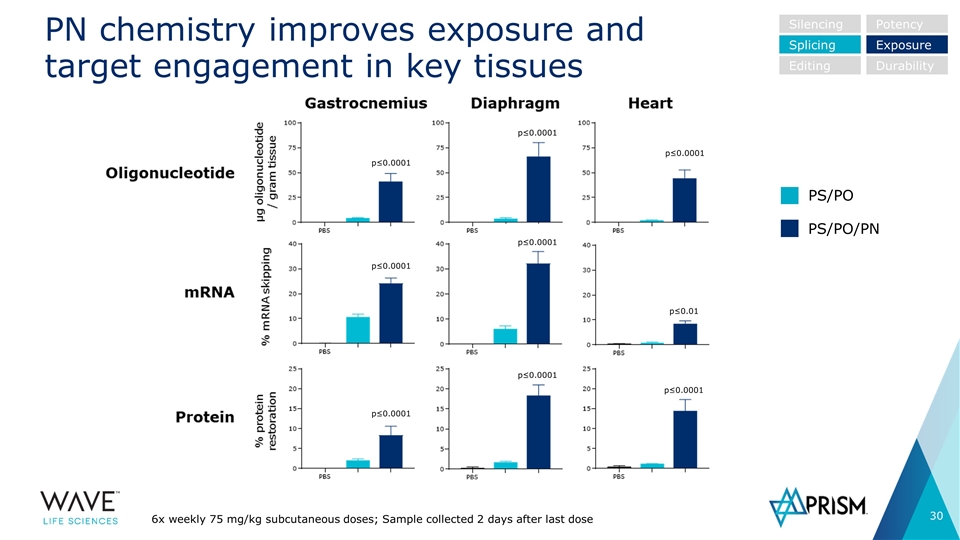
PN chemistry improves exposure and target engagement in key tissues Silencing Splicing Editing Potency Exposure Durability 6x weekly 75 mg/kg subcutaneous doses; Sample collected 2 days after last dose PS/PO PS/PO/PN p≤0.0001 p≤0.0001 p≤0.0001 p≤0.01 p≤0.0001 p≤0.0001 p≤0.0001 p≤0.0001 p≤0.0001
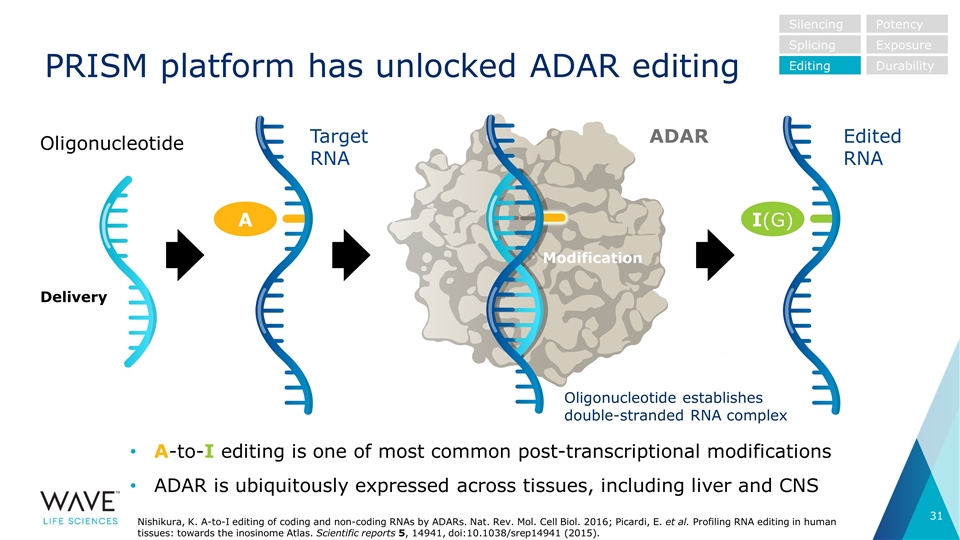
PRISM platform has unlocked ADAR editing A-to-I editing is one of most common post-transcriptional modifications ADAR is ubiquitously expressed across tissues, including liver and CNS ADAR Target RNA I(G) A Edited RNA Oligonucleotide establishes double-stranded RNA complex Oligonucleotide Modification Delivery Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016; Picardi, E. et al. Profiling RNA editing in human tissues: towards the inosinome Atlas. Scientific reports 5, 14941, doi:10.1038/srep14941 (2015). Silencing Splicing Editing Potency Exposure Durability
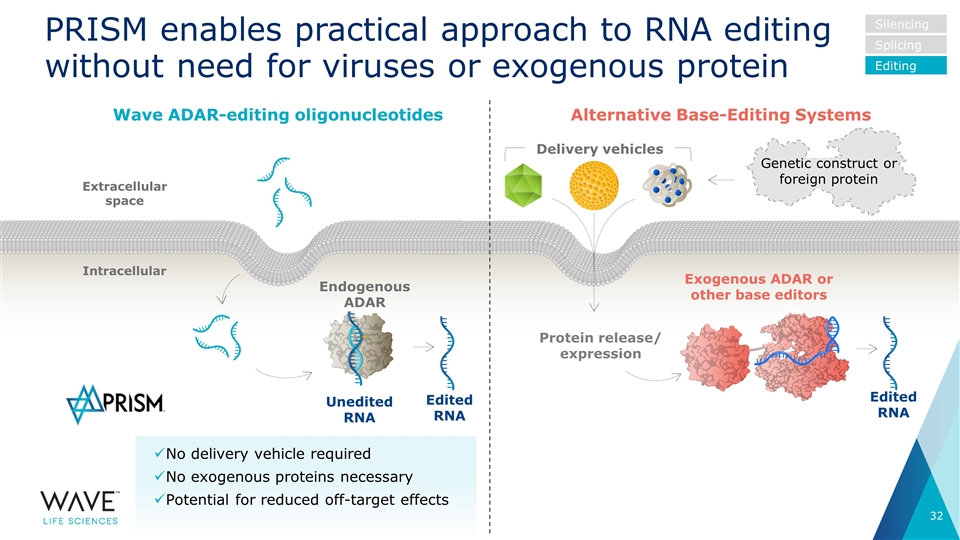
No delivery vehicle required No exogenous proteins necessary Potential for reduced off-target effects PRISM enables practical approach to RNA editing without need for viruses or exogenous protein Intracellular Extracellular space Endogenous ADAR Unedited RNA Wave ADAR-editing oligonucleotides Exogenous ADAR or other base editors Edited RNA Protein release/ expression Silencing Splicing Editing Delivery vehicles Alternative Base-Editing Systems Edited RNA Genetic construct or foreign protein
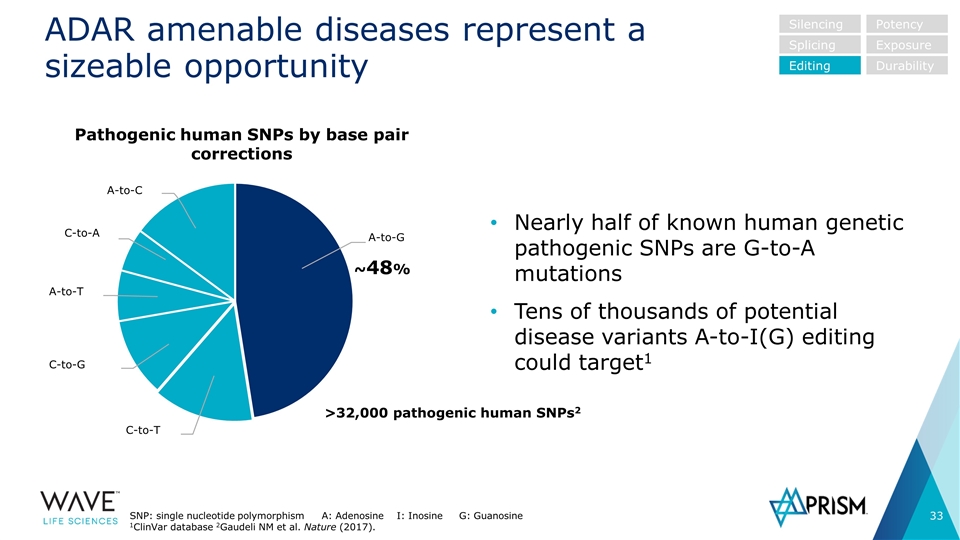
ADAR amenable diseases represent a sizeable opportunity Nearly half of known human genetic pathogenic SNPs are G-to-A mutations Tens of thousands of potential disease variants A-to-I(G) editing could target1 ~48% C-to-T C-to-G A-to-T C-to-A A-to-C A-to-G Pathogenic human SNPs by base pair corrections >32,000 pathogenic human SNPs2 Silencing Splicing Editing Potency Exposure Durability SNP: single nucleotide polymorphism A: Adenosine I: Inosine G: Guanosine 1ClinVar database 2Gaudeli NM et al. Nature (2017).
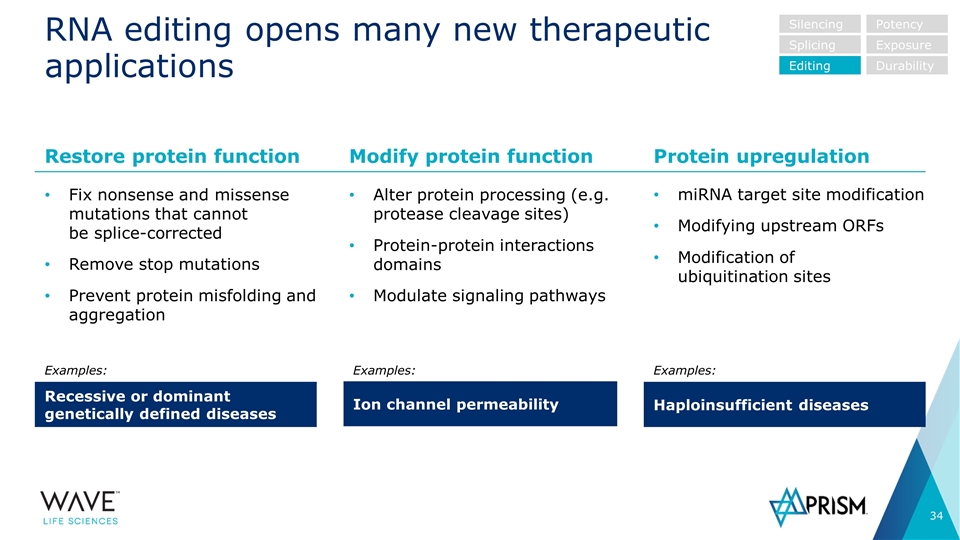
RNA editing opens many new therapeutic applications Fix nonsense and missense mutations that cannot be splice-corrected Remove stop mutations Prevent protein misfolding and aggregation Alter protein processing (e.g. protease cleavage sites) Protein-protein interactions domains Modulate signaling pathways miRNA target site modification Modifying upstream ORFs Modification of ubiquitination sites Restore protein function Recessive or dominant genetically defined diseases Modify protein function Ion channel permeability Protein upregulation Haploinsufficient diseases Examples: Examples: Examples: Silencing Splicing Editing Potency Exposure Durability
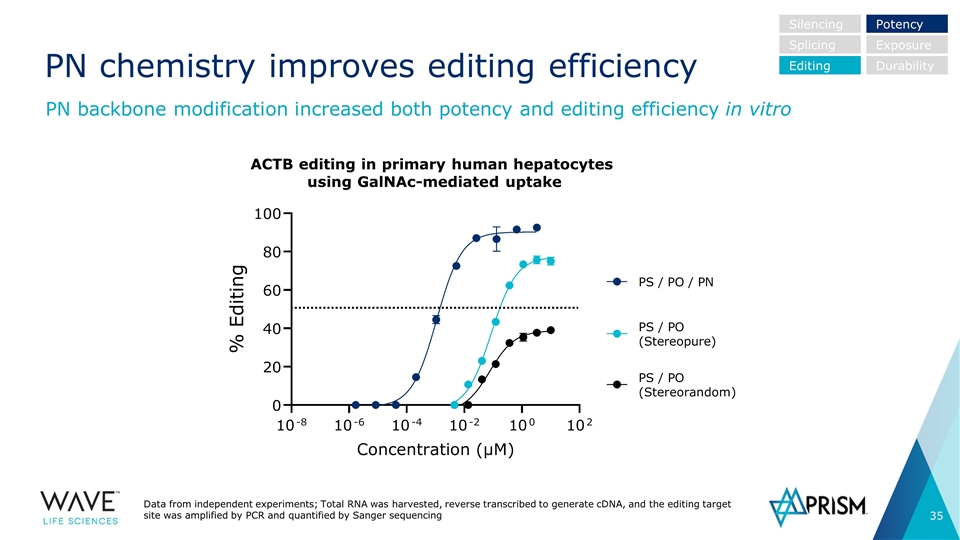
Data from independent experiments; Total RNA was harvested, reverse transcribed to generate cDNA, and the editing target site was amplified by PCR and quantified by Sanger sequencing PN chemistry improves editing efficiency PN backbone modification increased both potency and editing efficiency in vitro Silencing Splicing Editing Potency Exposure Durability ACTB editing in primary human hepatocytes using GalNAc-mediated uptake
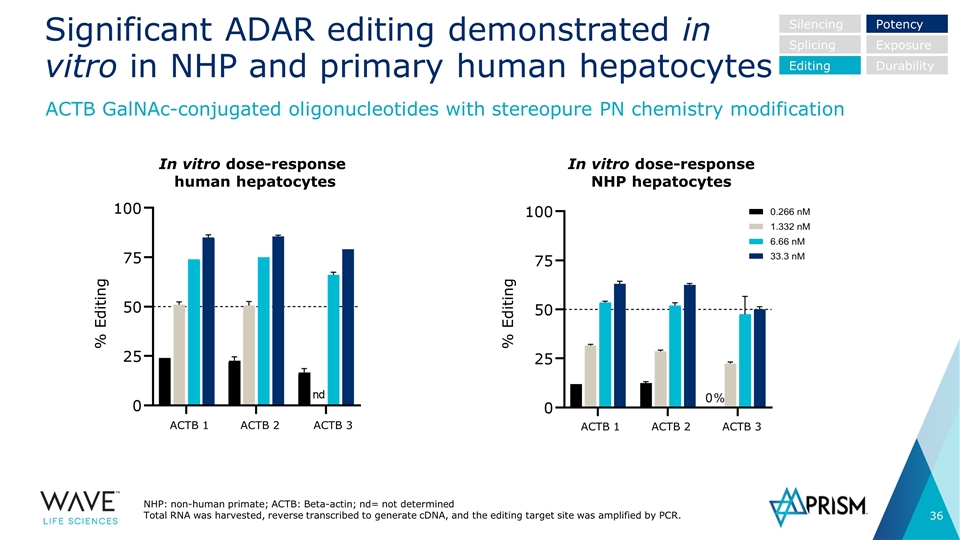
Significant ADAR editing demonstrated in vitro in NHP and primary human hepatocytes NHP: non-human primate; ACTB: Beta-actin; nd= not determined Total RNA was harvested, reverse transcribed to generate cDNA, and the editing target site was amplified by PCR. Silencing Splicing Editing Potency Exposure Durability In vitro dose-response human hepatocytes In vitro dose-response NHP hepatocytes % Editing % Editing ACTB 1 ACTB 2 ACTB 3 ACTB 1 ACTB 2 ACTB 3 ACTB GalNAc-conjugated oligonucleotides with stereopure PN chemistry modification
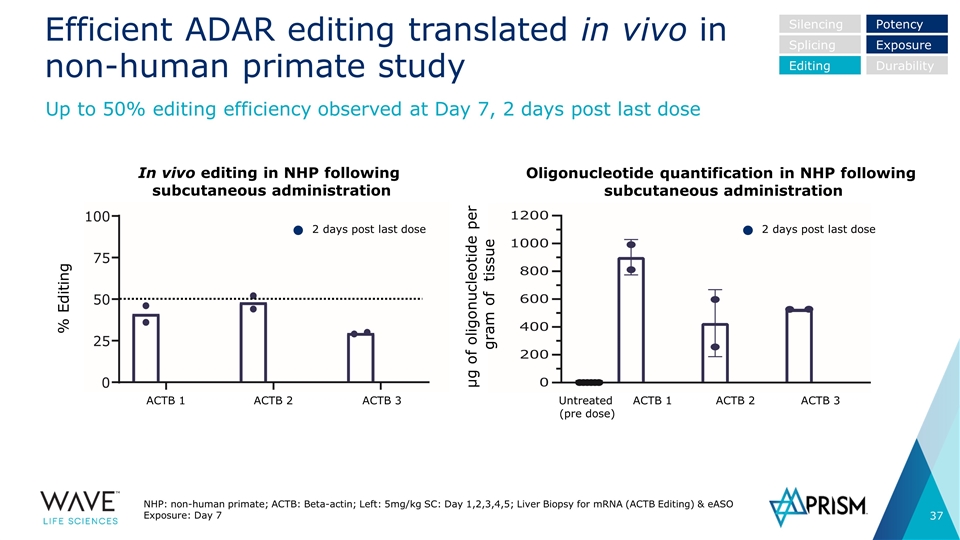
Efficient ADAR editing translated in vivo in non-human primate study NHP: non-human primate; ACTB: Beta-actin; Left: 5mg/kg SC: Day 1,2,3,4,5; Liver Biopsy for mRNA (ACTB Editing) & eASO Exposure: Day 7 Up to 50% editing efficiency observed at Day 7, 2 days post last dose Silencing Splicing Editing Potency Exposure Durability In vivo editing in NHP following subcutaneous administration Oligonucleotide quantification in NHP following subcutaneous administration % Editing µg of oligonucleotide per gram of tissue 2 days post last dose 2 days post last dose ACTB 1 ACTB 2 ACTB 3 ACTB 1 ACTB 2 ACTB 3 Untreated (pre dose)
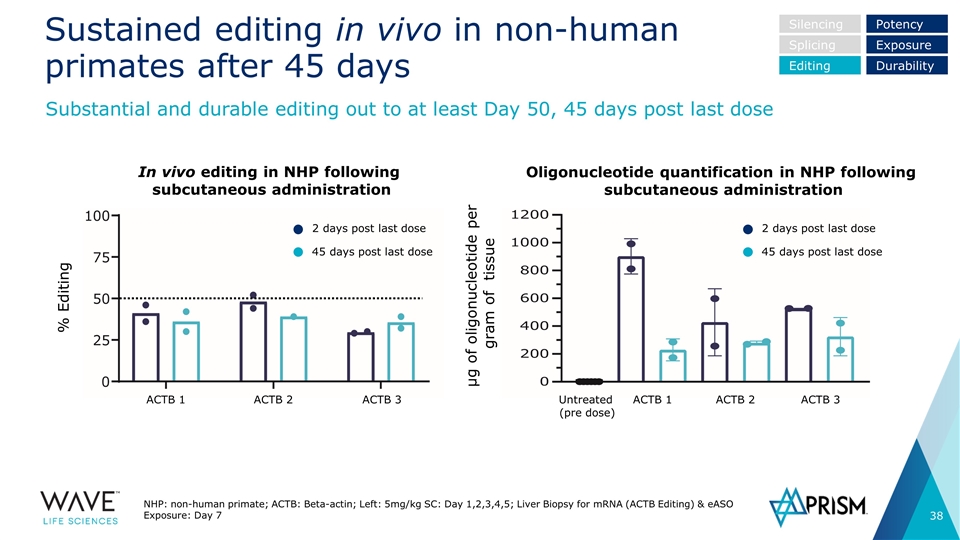
Sustained editing in vivo in non-human primates after 45 days NHP: non-human primate; ACTB: Beta-actin; Left: 5mg/kg SC: Day 1,2,3,4,5; Liver Biopsy for mRNA (ACTB Editing) & eASO Exposure: Day 7 Substantial and durable editing out to at least Day 50, 45 days post last dose Silencing Splicing Editing Potency Exposure Durability In vivo editing in NHP following subcutaneous administration Oligonucleotide quantification in NHP following subcutaneous administration 2 days post last dose 45 days post last dose 2 days post last dose 45 days post last dose ACTB 1 ACTB 2 ACTB 3 ACTB 1 ACTB 2 ACTB 3 Untreated (pre dose) % Editing µg of oligonucleotide per gram of tissue
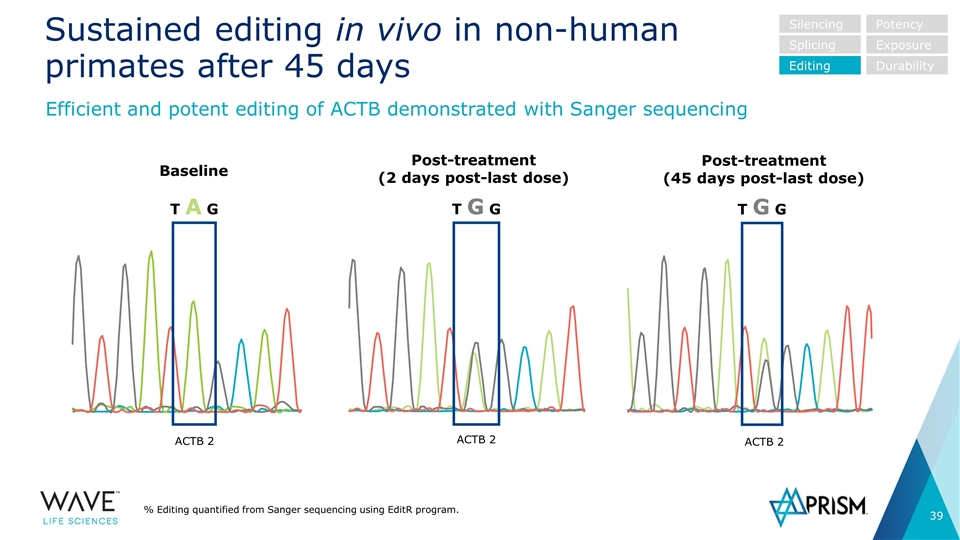
Sustained editing in vivo in non-human primates after 45 days % Editing quantified from Sanger sequencing using EditR program. Efficient and potent editing of ACTB demonstrated with Sanger sequencing Silencing Splicing Editing Potency Exposure Durability T A G T G G T G G Baseline Post-treatment (2 days post-last dose) Post-treatment (45 days post-last dose) ACTB 2 ACTB 2 ACTB 2
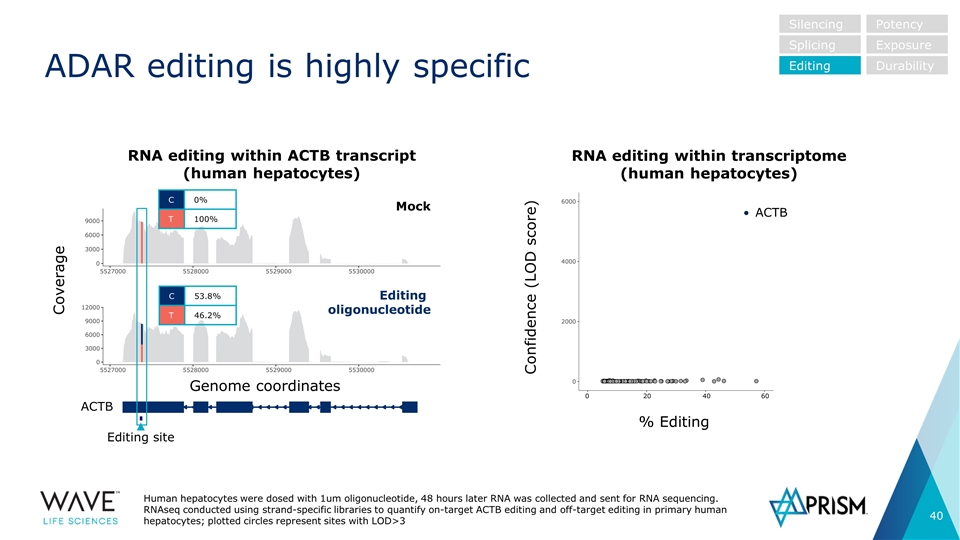
Editing site RNA editing within ACTB transcript (human hepatocytes) RNA editing within transcriptome (human hepatocytes) ADAR editing is highly specific Silencing Splicing Editing Potency Exposure Durability Coverage Genome coordinates ACTB C 0% T 100% C 53.8% T 46.2% ACTB Confidence (LOD score) % Editing Mock Editing oligonucleotide Human hepatocytes were dosed with 1um oligonucleotide, 48 hours later RNA was collected and sent for RNA sequencing. RNAseq conducted using strand-specific libraries to quantify on-target ACTB editing and off-target editing in primary human hepatocytes; plotted circles represent sites with LOD>3
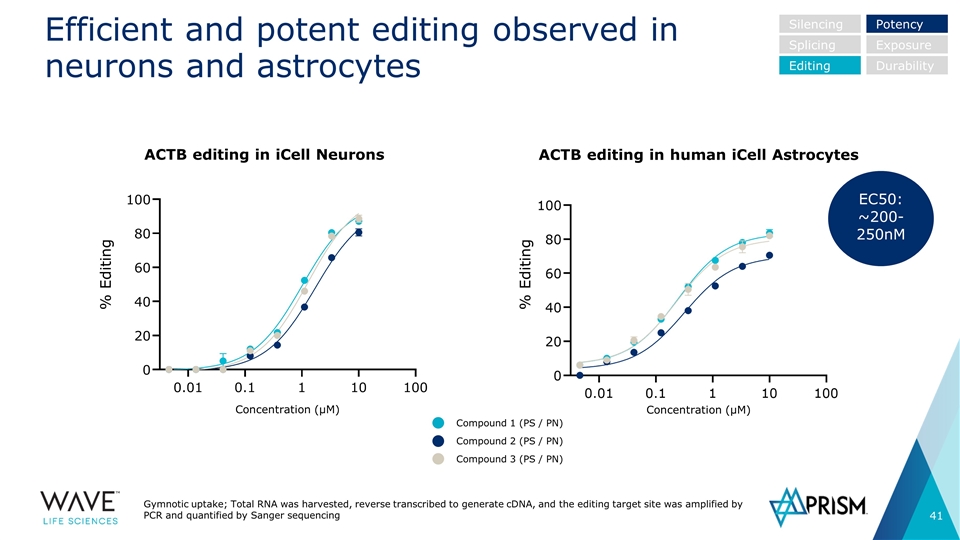
Efficient and potent editing observed in neurons and astrocytes Silencing Splicing Editing Potency Exposure Durability ACTB editing in iCell Neurons ACTB editing in human iCell Astrocytes Concentration (µM) % Editing % Editing Compound 2 (PS / PN) Compound 1 (PS / PN) Compound 3 (PS / PN) EC50: ~200-250nM Gymnotic uptake; Total RNA was harvested, reverse transcribed to generate cDNA, and the editing target site was amplified by PCR and quantified by Sanger sequencing Concentration (µM)
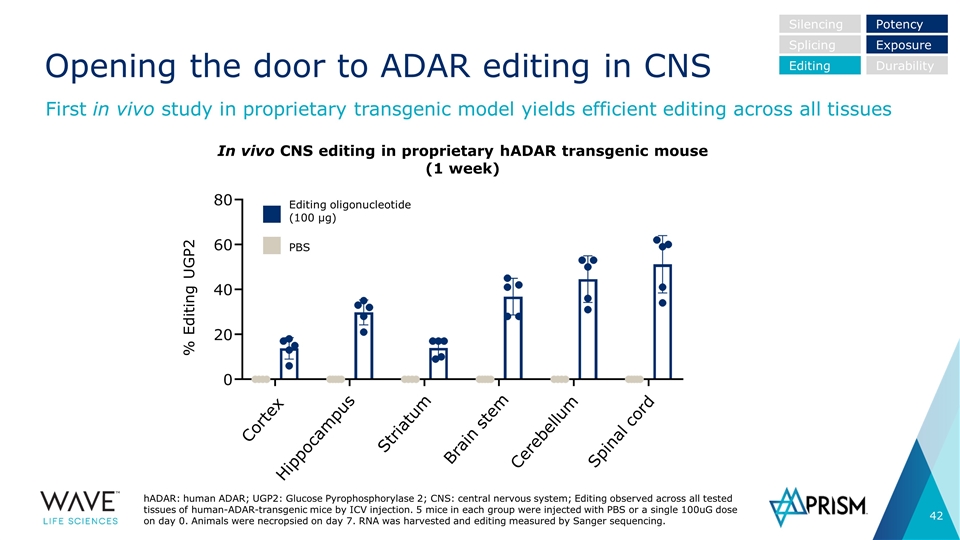
Opening the door to ADAR editing in CNS Silencing Splicing Editing Potency Exposure Durability In vivo CNS editing in proprietary hADAR transgenic mouse (1 week) Editing oligonucleotide (100 µg) PBS % Editing UGP2 Cortex Hippocampus Striatum Brain stem Cerebellum Spinal cord hADAR: human ADAR; UGP2: Glucose Pyrophosphorylase 2; CNS: central nervous system; Editing observed across all tested tissues of human-ADAR-transgenic mice by ICV injection. 5 mice in each group were injected with PBS or a single 100uG dose on day 0. Animals were necropsied on day 7. RNA was harvested and editing measured by Sanger sequencing. First in vivo study in proprietary transgenic model yields efficient editing across all tissues

Continued evolution of PRISM platform PN chemistry is a novel backbone chemistry modification Preclinical data demonstrates PN chemistry can enhance potency, durability and exposure across three modalities (silencing, splicing, editing) Platform innovations have unlocked ADAR editing modality Efficient and durable editing demonstrated in vivo in NHPs Expect to announce first ADAR editing program in a hepatic indication in 2020 Moving ADAR editing quickly into neurology Editing demonstrated in neurons and astrocytes in vitro and across CNS tissue types in vivo in first transgenic human ADAR mouse study Ongoing work to unlock new neurology targets with ADAR editing Sustained investment has yielded novel modality and pharmacology advances

Neurology pipeline & C9orf72 program Kenneth Rhodes, PhD SVP, Therapeutics Discovery

Focus on neurological diseases Neurological diseases represent one of the greatest medical challenges of our time PRISM can deliver oligonucleotide drug candidates that directly address the genetic drivers of neurologic diseases PRISM-derived oligonucleotides access neurons and glia without need for transfection, encapsulation or other modifications The range of modalities offered by PRISM (silencing, splicing and ADAR editing), unlocks therapeutic opportunities in a broad range of neurological diseases
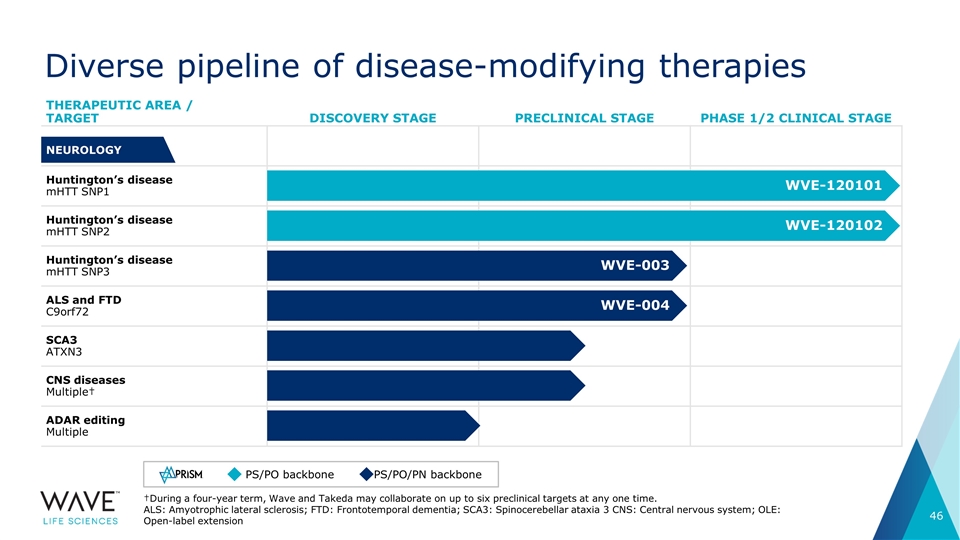
Diverse pipeline of disease-modifying therapies THERAPEUTIC AREA / TARGET DISCOVERY STAGE PRECLINICAL STAGE PHASE 1/2 CLINICAL STAGE Huntington’s disease mHTT SNP1 Huntington’s disease mHTT SNP2 Huntington’s disease mHTT SNP3 ALS and FTD C9orf72 SCA3 ATXN3 CNS diseases Multiple† ADAR editing Multiple WVE-120101 PS/PO backbone PS/PO/PN backbone †During a four-year term, Wave and Takeda may collaborate on up to six preclinical targets at any one time. ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia; SCA3: Spinocerebellar ataxia 3 CNS: Central nervous system; OLE: Open-label extension NEUROLOGY WVE-120102 WVE-003 WVE-004
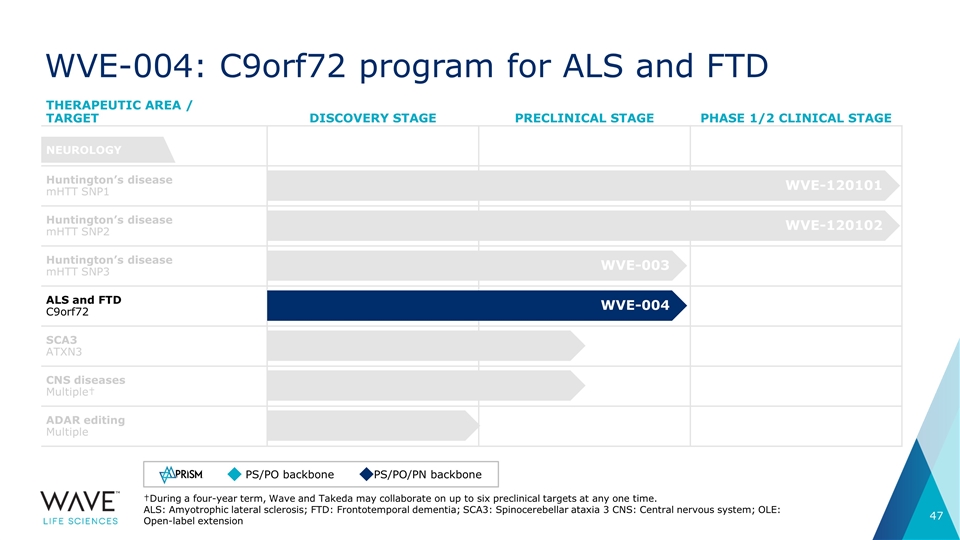
WVE-004: C9orf72 program for ALS and FTD THERAPEUTIC AREA / TARGET DISCOVERY STAGE PRECLINICAL STAGE PHASE 1/2 CLINICAL STAGE Huntington’s disease mHTT SNP1 Huntington’s disease mHTT SNP2 Huntington’s disease mHTT SNP3 ALS and FTD C9orf72 SCA3 ATXN3 CNS diseases Multiple† ADAR editing Multiple NEUROLOGY †During a four-year term, Wave and Takeda may collaborate on up to six preclinical targets at any one time. ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia; SCA3: Spinocerebellar ataxia 3 CNS: Central nervous system; OLE: Open-label extension WVE-120101 WVE-120102 WVE-003 WVE-004 PS/PO backbone PS/PO/PN backbone
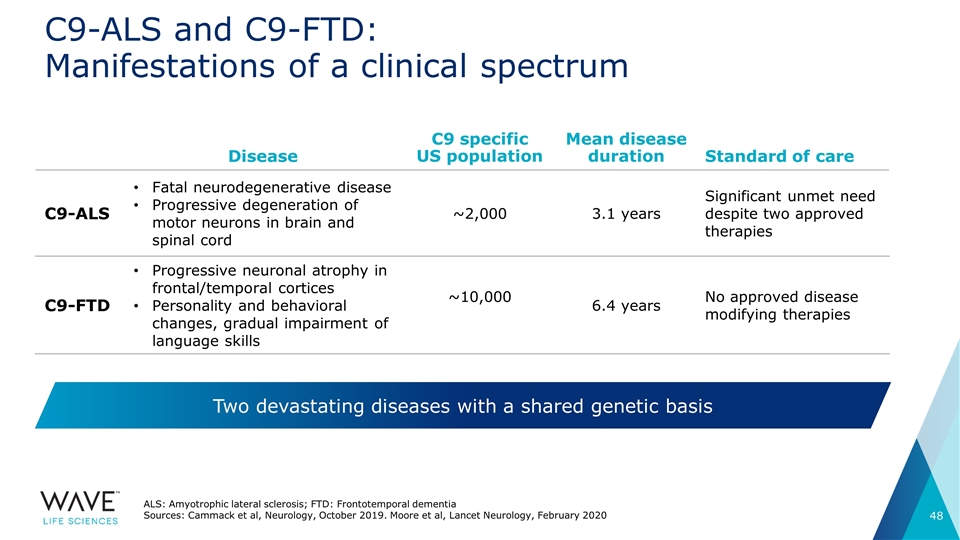
C9-ALS and C9-FTD: Manifestations of a clinical spectrum Disease C9 specific US population Mean disease duration Standard of care C9-ALS Fatal neurodegenerative disease Progressive degeneration of motor neurons in brain and spinal cord ~2,000 3.1 years Significant unmet need despite two approved therapies C9-FTD Progressive neuronal atrophy in frontal/temporal cortices Personality and behavioral changes, gradual impairment of language skills ~10,000 6.4 years No approved disease modifying therapies Two devastating diseases with a shared genetic basis ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia Sources: Cammack et al, Neurology, October 2019. Moore et al, Lancet Neurology, February 2020
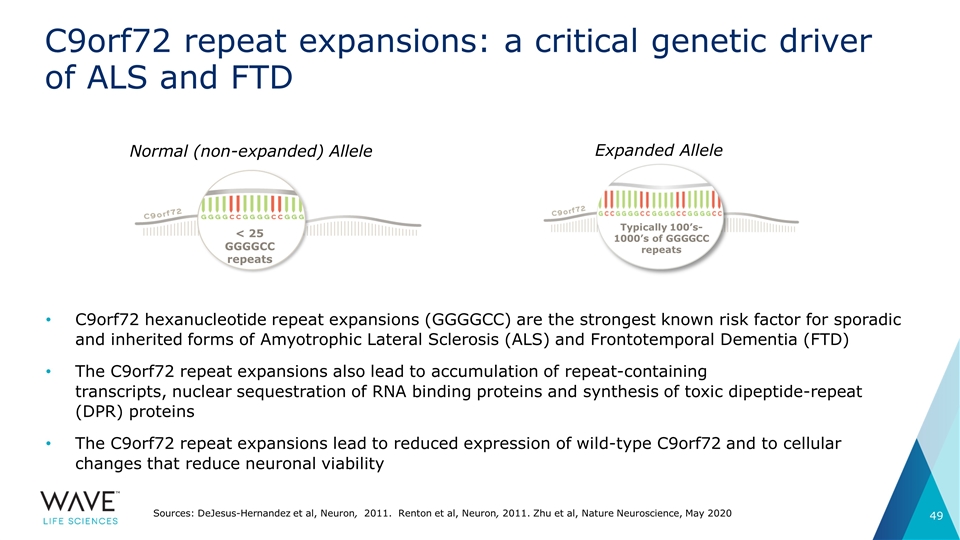
C9orf72 repeat expansions: a critical genetic driver of ALS and FTD Normal (non-expanded) Allele < 25 GGGGCC repeats Expanded Allele Sources: DeJesus-Hernandez et al, Neuron, 2011. Renton et al, Neuron, 2011. Zhu et al, Nature Neuroscience, May 2020 Typically 100’s-1000’s of GGGGCC repeats C9orf72 hexanucleotide repeat expansions (GGGGCC) are the strongest known risk factor for sporadic and inherited forms of Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal Dementia (FTD) The C9orf72 repeat expansions also lead to accumulation of repeat-containing transcripts, nuclear sequestration of RNA binding proteins and synthesis of toxic dipeptide-repeat (DPR) proteins The C9orf72 repeat expansions lead to reduced expression of wild-type C9orf72 and to cellular changes that reduce neuronal viability
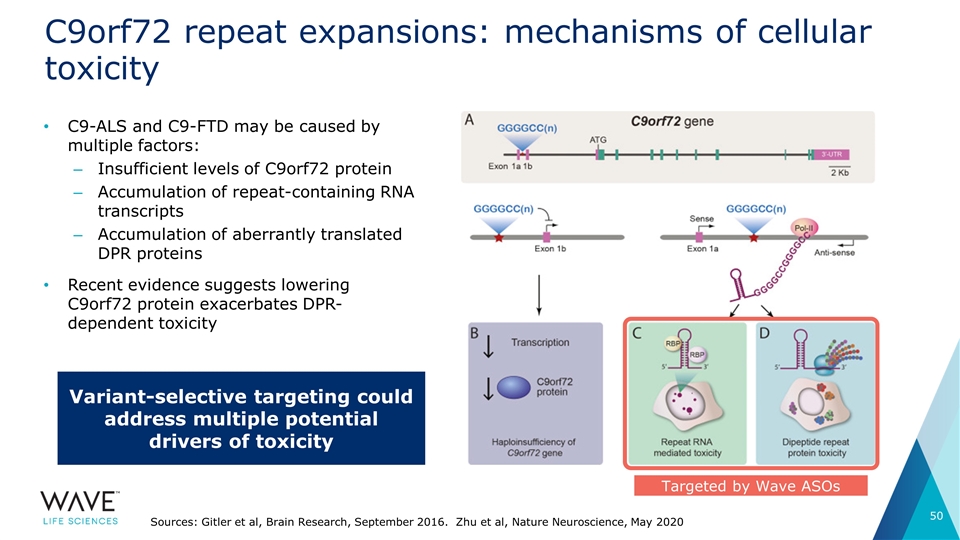
C9orf72 repeat expansions: mechanisms of cellular toxicity C9-ALS and C9-FTD may be caused by multiple factors: Insufficient levels of C9orf72 protein Accumulation of repeat-containing RNA transcripts Accumulation of aberrantly translated DPR proteins Recent evidence suggests lowering C9orf72 protein exacerbates DPR-dependent toxicity Sources: Gitler et al, Brain Research, September 2016. Zhu et al, Nature Neuroscience, May 2020 Targeted by Wave ASOs Variant-selective targeting could address multiple potential drivers of toxicity
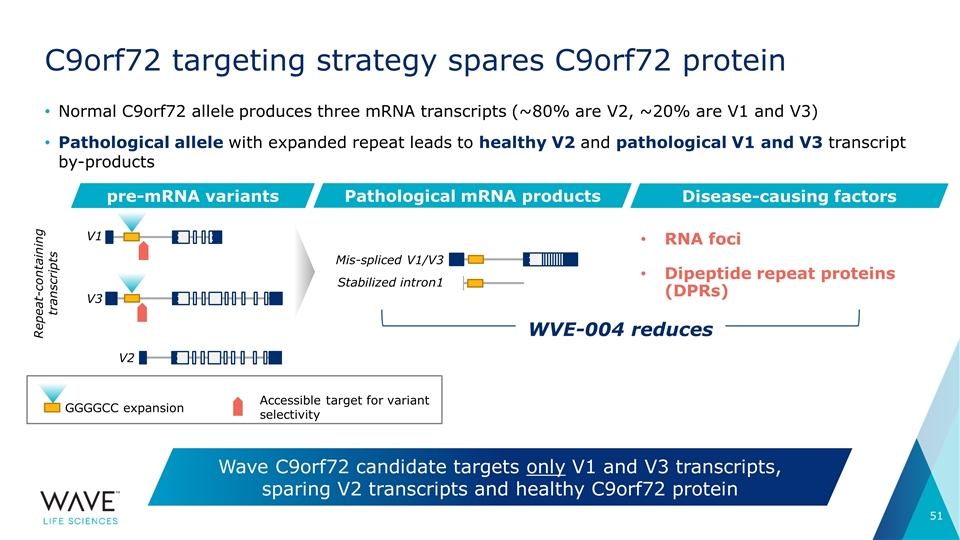
Normal C9orf72 allele produces three mRNA transcripts (~80% are V2, ~20% are V1 and V3) Pathological allele with expanded repeat leads to healthy V2 and pathological V1 and V3 transcript by-products C9orf72 targeting strategy spares C9orf72 protein Wave C9orf72 candidate targets only V1 and V3 transcripts, sparing V2 transcripts and healthy C9orf72 protein pre-mRNA variants Pathological mRNA products V1 V2 Mis-spliced V1/V3 Stabilized intron1 V3 Disease-causing factors RNA foci Dipeptide repeat proteins (DPRs) GGGGCC expansion Accessible target for variant selectivity WVE-004 reduces Repeat-containing transcripts
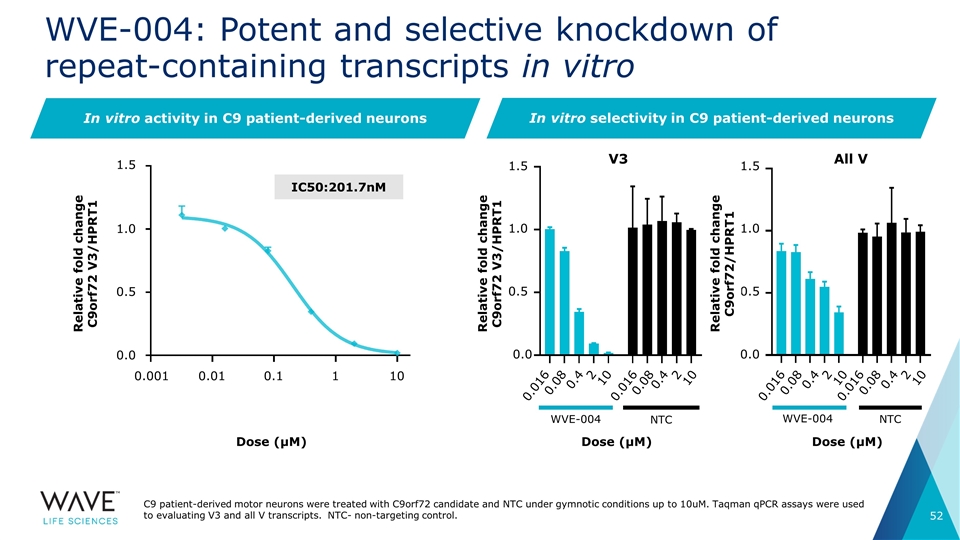
WVE-004: Potent and selective knockdown of repeat-containing transcripts in vitro V3 Dose (μM) All V WVE-004 NTC Dose (μM) In vitro activity in C9 patient-derived neurons WVE-004 NTC Dose (μM) IC50:201.7nM In vitro selectivity in C9 patient-derived neurons C9 patient-derived motor neurons were treated with C9orf72 candidate and NTC under gymnotic conditions up to 10uM. Taqman qPCR assays were used to evaluating V3 and all V transcripts. NTC- non-targeting control. Relative fold change C9orf72 V3/HPRT1 1.5 1.0 0.5 0.0 0.001 0.01 0.1 1 10 Relative fold change C9orf72 V3/HPRT1 0.016 0.08 0.4 2 10 0.016 0.08 0.4 2 10 0.016 0.08 0.4 2 10 0.016 0.08 0.4 2 10 1.5 1.0 0.5 0.0 1.5 1.0 0.5 0.0 Relative fold change C9orf72/HPRT1
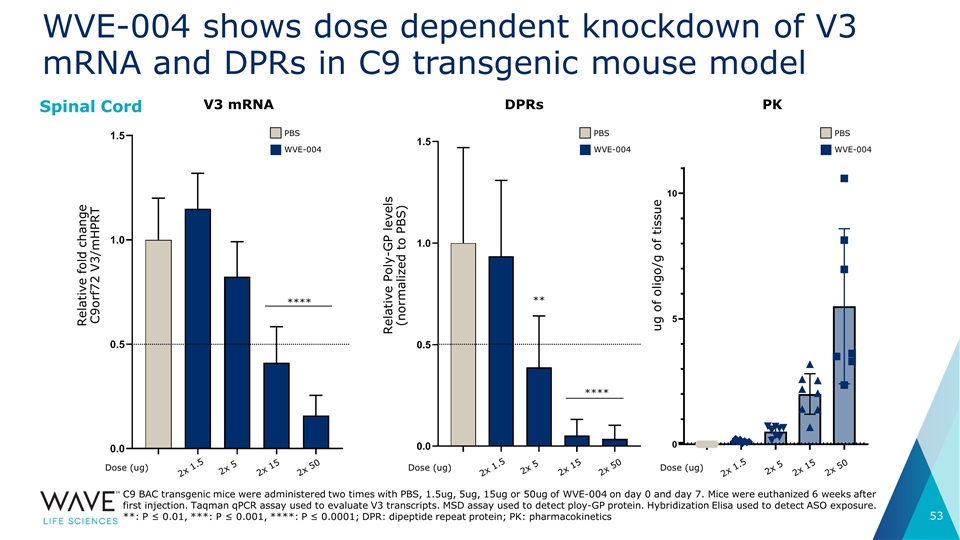
WVE-004 shows dose dependent knockdown of V3 mRNA and DPRs in C9 transgenic mouse model Spinal Cord C9 BAC transgenic mice were administered two times with PBS, 1.5ug, 5ug, 15ug or 50ug of WVE-004 on day 0 and day 7. Mice were euthanized 6 weeks after first injection. Taqman qPCR assay used to evaluate V3 transcripts. MSD assay used to detect ploy-GP protein. Hybridization Elisa used to detect ASO exposure. **: P ≤ 0.01, ***: P ≤ 0.001, ****: P ≤ 0.0001; DPR: dipeptide repeat protein; PK: pharmacokinetics V3 mRNA **** Dose (ug) 2x 1.5 2x 5 2x 15 2x 50 2x 1.5 2x 5 2x 15 2x 50 PBS WVE-004 Relative fold change C9orf72 V3/mHPRT DPRs **** ** Relative Poly-GP levels (normalized to PBS) PBS WVE-004 ug of oligo/g of tissue PK PBS WVE-004 Dose (ug) Dose (ug) 2x 1.5 2x 5 2x 15 2x 50
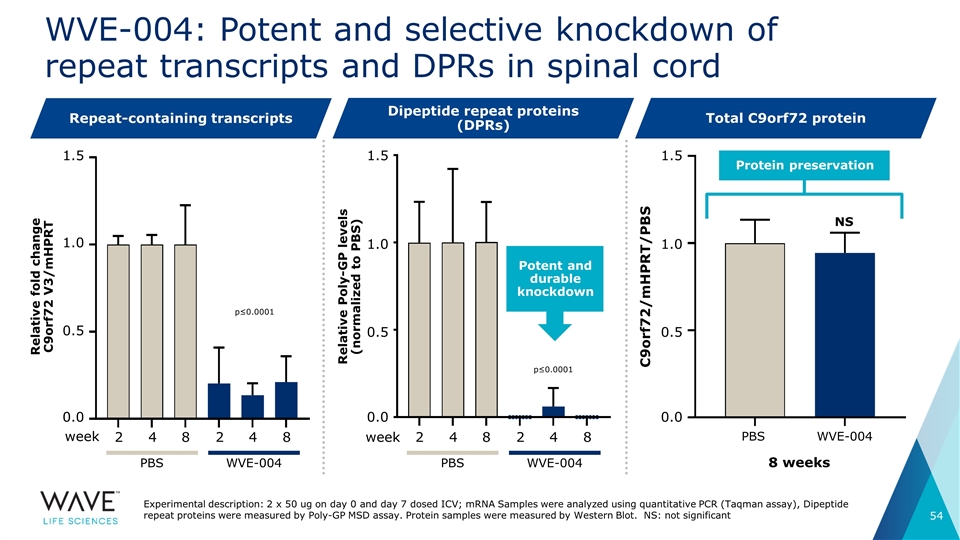
WVE-004: Potent and selective knockdown of repeat transcripts and DPRs in spinal cord 2 4 8 week 2 4 8 1.5 WVE-004 PBS 2 4 8 week 2 4 8 WVE-004 PBS Potent and durable knockdown 8 weeks WVE-004 PBS NS Repeat-containing transcripts Dipeptide repeat proteins (DPRs) Total C9orf72 protein Experimental description: 2 x 50 ug on day 0 and day 7 dosed ICV; mRNA Samples were analyzed using quantitative PCR (Taqman assay), Dipeptide repeat proteins were measured by Poly-GP MSD assay. Protein samples were measured by Western Blot. NS: not significant Protein preservation 1.0 0.5 0.0 1.5 1.0 0.5 0.0 Relative fold change C9orf72 V3/mHPRT Relative Poly-GP levels (normalized to PBS) 1.5 1.0 0.5 0.0 p≤0.0001 p≤0.0001 C9orf72/mHPRT/PBS
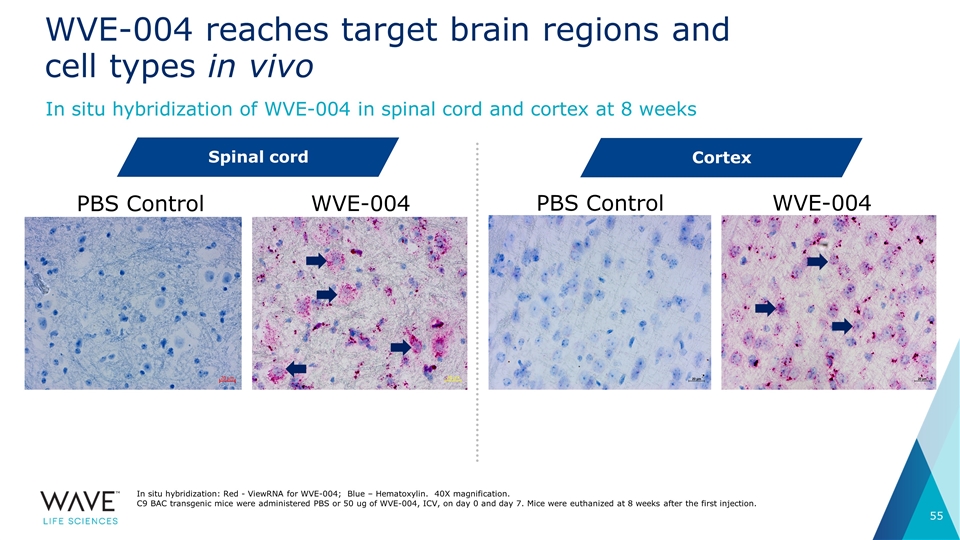
WVE-004 reaches target brain regions and cell types in vivo In situ hybridization: Red - ViewRNA for WVE-004; Blue – Hematoxylin. 40X magnification. C9 BAC transgenic mice were administered PBS or 50 ug of WVE-004, ICV, on day 0 and day 7. Mice were euthanized at 8 weeks after the first injection. Spinal cord PBS Control WVE-004 PBS Control WVE-004 Cortex In situ hybridization of WVE-004 in spinal cord and cortex at 8 weeks
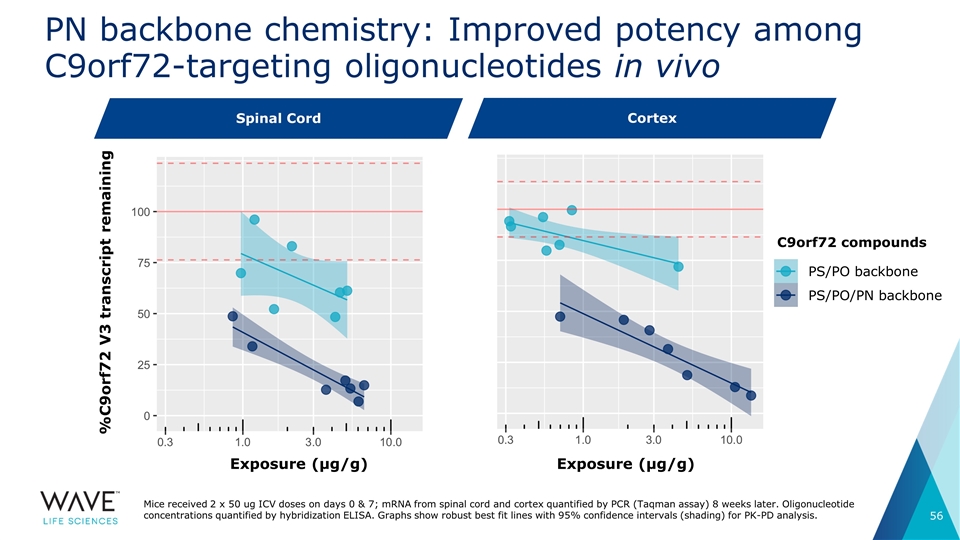
PN backbone chemistry: Improved potency among C9orf72-targeting oligonucleotides in vivo Exposure (µg/g) Exposure (µg/g) C9orf72 compounds Spinal cord Cortex PS/PO backbone PS/PO/PN backbone %C9orf72 V3 transcript remaining Mice received 2 x 50 ug ICV doses on days 0 & 7; mRNA from spinal cord and cortex quantified by PCR (Taqman assay) 8 weeks later. Oligonucleotide concentrations quantified by hybridization ELISA. Graphs show robust best fit lines with 95% confidence intervals (shading) for PK-PD analysis. Spinal Cord
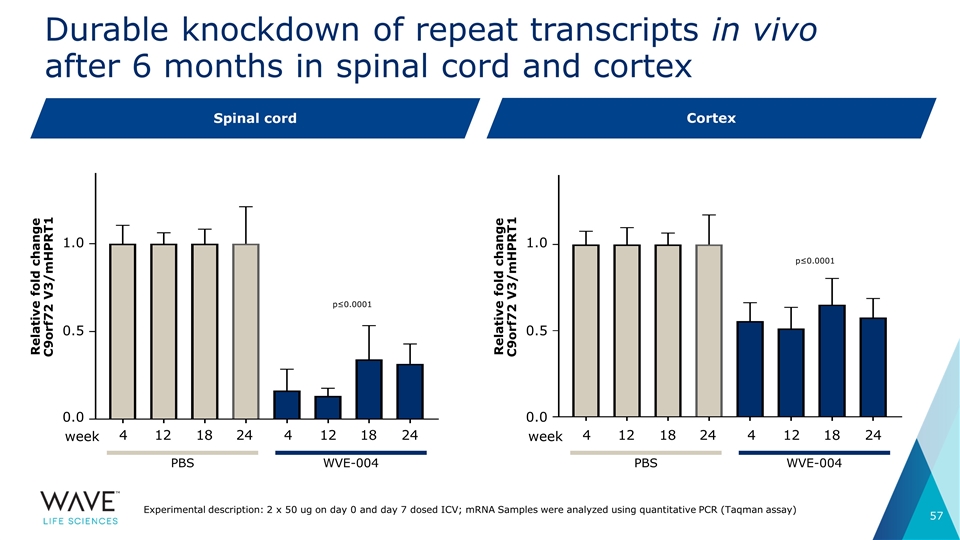
4 12 18 12 18 24 4 24 Durable knockdown of repeat transcripts in vivo after 6 months in spinal cord and cortex Spinal cord Cortex 1.0 0.5 0.0 Relative fold change C9orf72 V3/mHPRT1 WVE-004 PBS week 4 12 18 12 18 24 4 24 1.0 0.5 0.0 Relative fold change C9orf72 V3/mHPRT1 WVE-004 PBS week Experimental description: 2 x 50 ug on day 0 and day 7 dosed ICV; mRNA Samples were analyzed using quantitative PCR (Taqman assay) p≤0.0001 p≤0.0001
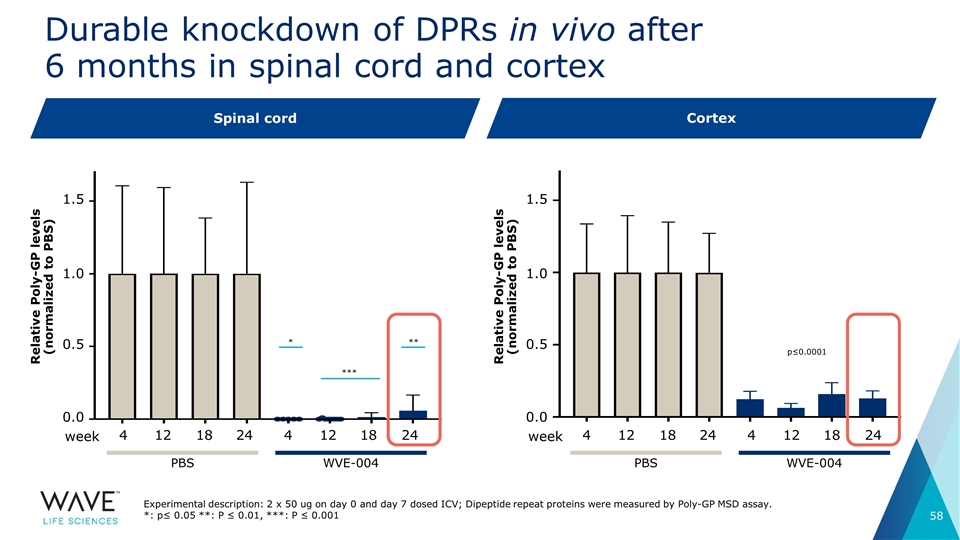
Durable knockdown of DPRs in vivo after 6 months in spinal cord and cortex 4 12 18 12 18 24 4 24 Spinal cord Cortex 1.5 0.5 0.0 WVE-004 PBS week 4 12 18 12 18 24 4 24 WVE-004 PBS week 1.0 Relative Poly-GP levels (normalized to PBS) 1.5 0.5 0.0 1.0 Experimental description: 2 x 50 ug on day 0 and day 7 dosed ICV; Dipeptide repeat proteins were measured by Poly-GP MSD assay. *: p≤ 0.05 **: P ≤ 0.01, ***: P ≤ 0.001 p≤0.0001 * *** ** Relative Poly-GP levels (normalized to PBS)
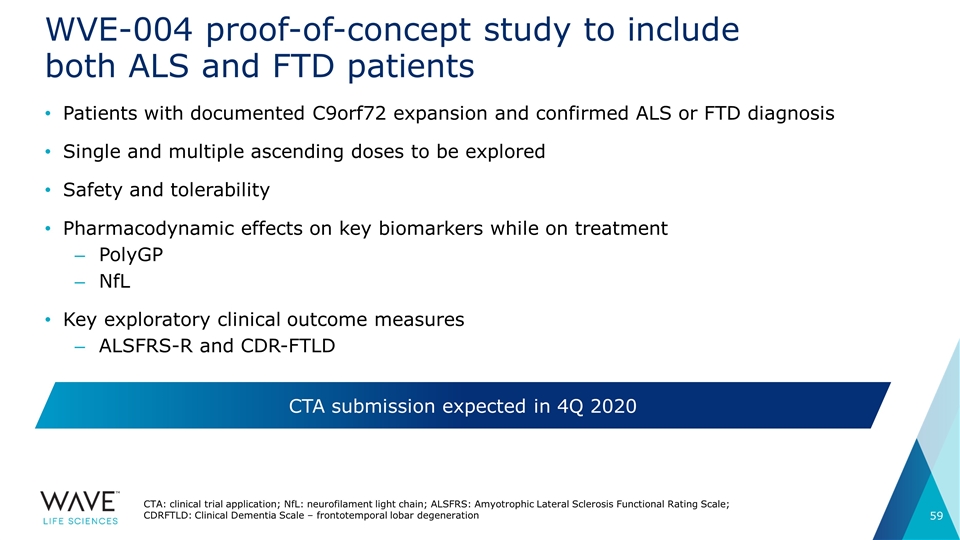
WVE-004 proof-of-concept study to include both ALS and FTD patients Patients with documented C9orf72 expansion and confirmed ALS or FTD diagnosis Single and multiple ascending doses to be explored Safety and tolerability Pharmacodynamic effects on key biomarkers while on treatment PolyGP NfL Key exploratory clinical outcome measures ALSFRS-R and CDR-FTLD CTA submission expected in 4Q 2020 CTA: clinical trial application; NfL: neurofilament light chain; ALSFRS: Amyotrophic Lateral Sclerosis Functional Rating Scale; CDRFTLD: Clinical Dementia Scale – frontotemporal lobar degeneration
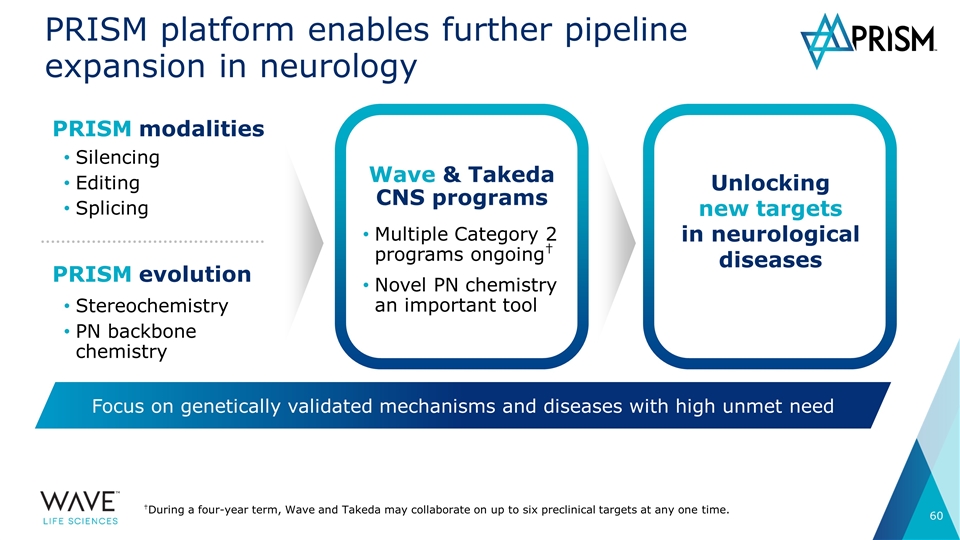
PRISM platform enables further pipeline expansion in neurology PRISM modalities Silencing Editing Splicing PRISM evolution Stereochemistry PN backbone chemistry Unlocking new targets in neurological diseases Wave & Takeda CNS programs Multiple Category 2 programs ongoing† Novel PN chemistry an important tool Focus on genetically validated mechanisms and diseases with high unmet need †During a four-year term, Wave and Takeda may collaborate on up to six preclinical targets at any one time.

Conclusion Paul Bolno, MD, MBA President and CEO
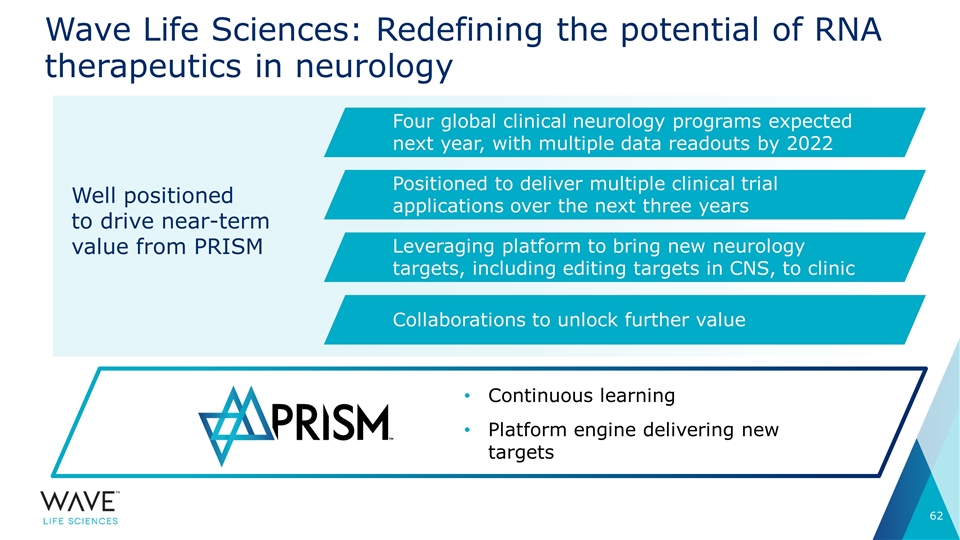
Wave Life Sciences: Redefining the potential of RNA therapeutics in neurology Continuous learning Platform engine delivering new targets Four global clinical neurology programs expected next year, with multiple data readouts by 2022 Positioned to deliver multiple clinical trial applications over the next three years Leveraging platform to bring new neurology targets, including editing targets in CNS, to clinic Collaborations to unlock further value Well positioned to drive near-term value from PRISM

Q&A

Analyst & Investor Research Webcast August 25, 2020































































