
Wave Life Sciences Corporate Presentation June 2, 2021 Exhibit 99.2

Forward-looking statements This document contains forward-looking statements. All statements other than statements of historical facts contained in this document, including statements regarding possible or assumed future results of operations, preclinical and clinical studies, business strategies, research and development plans, collaborations and partnerships, regulatory activities and timing thereof, competitive position, potential growth opportunities, use of proceeds and the effects of competition are forward-looking statements. These statements involve known and unknown risks, uncertainties and other important factors that may cause the actual results, performance or achievements of Wave Life Sciences Ltd. (the “Company”) to be materially different from any future results, performance or achievements expressed or implied by the forward-looking statements. In some cases, you can identify forward-looking statements by terms such as “may,” “will,” “should,” “expect,” “plan,” “aim,” “anticipate,” “could,” “intend,” “target,” “project,” “contemplate,” “believe,” “estimate,” “predict,” “potential” or “continue” or the negative of these terms or other similar expressions. The forward-looking statements in this presentation are only predictions. The Company has based these forward-looking statements largely on its current expectations and projections about future events and financial trends that it believes may affect the Company’s business, financial condition and results of operations. These forward-looking statements speak only as of the date of this presentation and are subject to a number of risks, uncertainties and assumptions, including those listed under Risk Factors in the Company’s Form 10-K and other filings with the SEC, some of which cannot be predicted or quantified and some of which are beyond the Company’s control. The events and circumstances reflected in the Company’s forward-looking statements may not be achieved or occur, and actual results could differ materially from those projected in the forward-looking statements. Moreover, the Company operates in a dynamic industry and economy. New risk factors and uncertainties may emerge from time to time, and it is not possible for management to predict all risk factors and uncertainties that the Company may face. Except as required by applicable law, the Company does not plan to publicly update or revise any forward-looking statements contained herein, whether as a result of any new information, future events, changed circumstances or otherwise.
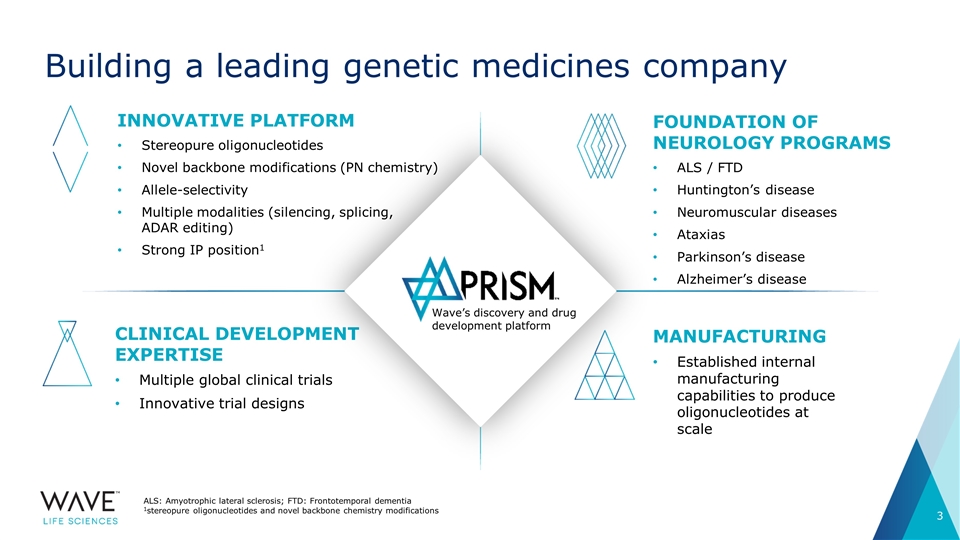
Building a leading genetic medicines company ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia 1stereopure oligonucleotides and novel backbone chemistry modifications Innovative platform Stereopure oligonucleotides Novel backbone modifications (PN chemistry) Allele-selectivity Multiple modalities (silencing, splicing, ADAR editing) Strong IP position1 Foundation of NEUROLOGY programs ALS / FTD Huntington’s disease Neuromuscular diseases Ataxias Parkinson’s disease Alzheimer’s disease Clinical development expertise Multiple global clinical trials Innovative trial designs Manufacturing Established internal manufacturing capabilities to produce oligonucleotides at scale Wave’s discovery and drug development platform
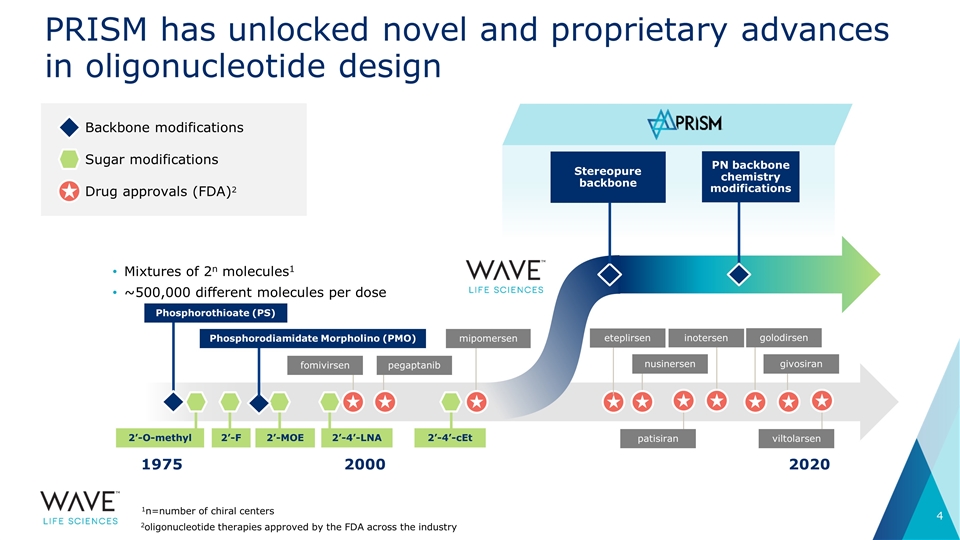
PRISM has unlocked novel and proprietary advances in oligonucleotide design Backbone modifications Sugar modifications Drug approvals (FDA)2 1975 2020 2000 Mixtures of 2n molecules1 ~500,000 different molecules per dose fomivirsen pegaptanib Phosphorothioate (PS) mipomersen nusinersen PN backbone chemistry modifications Stereopure backbone 2’-4’-cEt 2’-O-methyl 2’-F 2’-4’-LNA 1n=number of chiral centers 2’-MOE Phosphorodiamidate Morpholino (PMO) eteplirsen golodirsen givosiran patisiran inotersen viltolarsen 2oligonucleotide therapies approved by the FDA across the industry
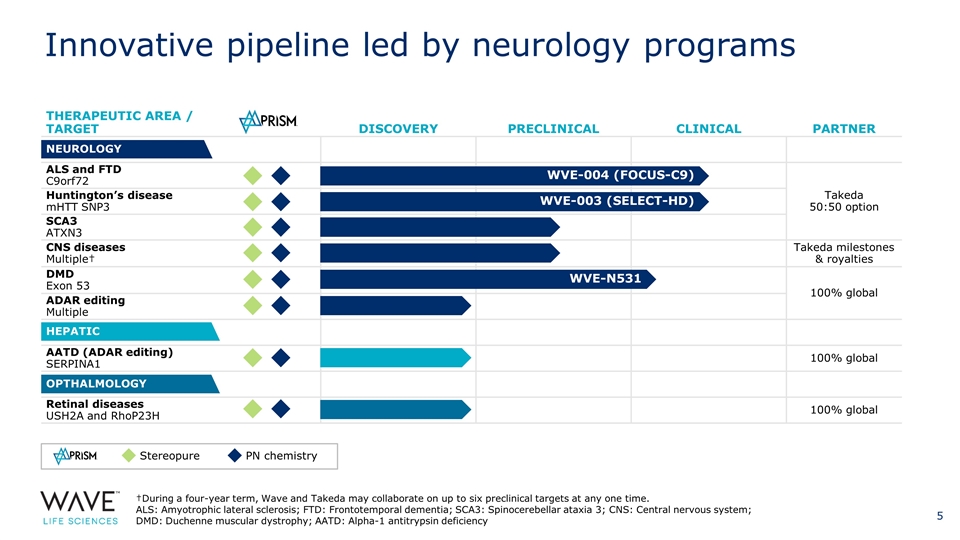
THERAPEUTIC AREA / TARGET DISCOVERY PRECLINICAL CLINICAL PARTNER ALS and FTD C9orf72 Takeda 50:50 option Huntington’s disease mHTT SNP3 SCA3 ATXN3 CNS diseases Multiple† Takeda milestones & royalties DMD Exon 53 100% global ADAR editing Multiple AATD (ADAR editing) SERPINA1 100% global Retinal diseases USH2A and RhoP23H 100% global NEUROLOGY HEPATIC OPTHALMOLOGY WVE-004 (FOCUS-C9) WVE-003 (SELECT-HD) †During a four-year term, Wave and Takeda may collaborate on up to six preclinical targets at any one time. ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia; SCA3: Spinocerebellar ataxia 3; CNS: Central nervous system; DMD: Duchenne muscular dystrophy; AATD: Alpha-1 antitrypsin deficiency Stereopure PN chemistry WVE-N531 Innovative pipeline led by neurology programs
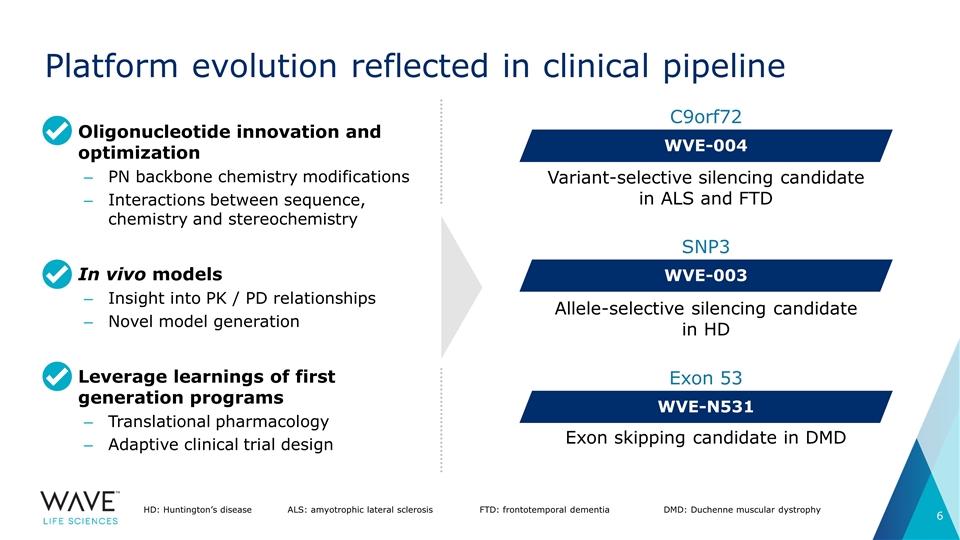
Platform evolution reflected in clinical pipeline Variant-selective silencing candidate in ALS and FTD WVE-004 C9orf72 WVE-003 SNP3 Allele-selective silencing candidate in HD WVE-N531 Exon 53 Exon skipping candidate in DMD HD: Huntington’s diseaseALS: amyotrophic lateral sclerosisFTD: frontotemporal dementia DMD: Duchenne muscular dystrophy Oligonucleotide innovation and optimization PN backbone chemistry modifications Interactions between sequence, chemistry and stereochemistry In vivo models Insight into PK / PD relationships Novel model generation Leverage learnings of first generation programs Translational pharmacology Adaptive clinical trial design

WVE-004 Amyotrophic Lateral Sclerosis (ALS) Frontotemporal Dementia (FTD)
C9orf72 repeat expansions: One of the most common genetic causes of ALS and FTD Typically 100’s-1000’s of GGGGCC repeats Amyotrophic Lateral Sclerosis (ALS) Frontotemporal Dementia (FTD) Hexanucleotide (G4C2)- repeat expansions in C9orf72 gene are common autosomal dominate cause for ALS and FTD Different manifestations across a clinical spectrum Fatal neurodegenerative disease Progressive degeneration of motor neurons in brain and spinal cord C9-specific ALS: ~2,000 patients in US Progressive neuronal degeneration in frontal/temporal cortices Personality and behavioral changes, gradual impairment of language skills C9-specific FTD: ~10,000 patients in US WVE-004 is the first therapy in clinical development for both C9-ALS and C9-FTD Sources: Balendra et al, EMBO Mol Med, 2017; Brown et al, NEJM, 2017, DeJesus-Hernandez et al, Neuron, 2011. Renton et al, Neuron, 2011. Zhu et al, Nature Neuroscience, May 2020, Stevens et al, Neurology 1998 Neuro C9orf72
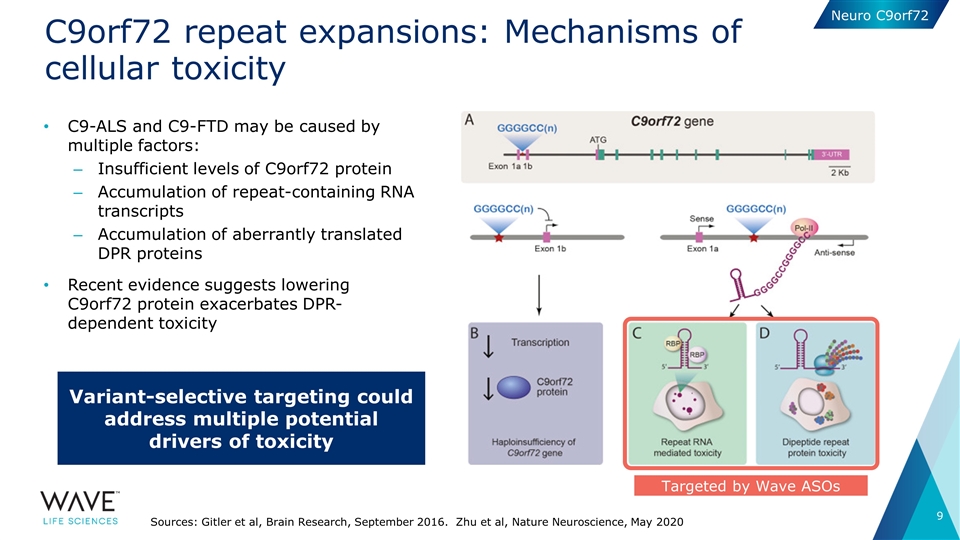
C9orf72 repeat expansions: Mechanisms of cellular toxicity C9-ALS and C9-FTD may be caused by multiple factors: Insufficient levels of C9orf72 protein Accumulation of repeat-containing RNA transcripts Accumulation of aberrantly translated DPR proteins Recent evidence suggests lowering C9orf72 protein exacerbates DPR-dependent toxicity Sources: Gitler et al, Brain Research, September 2016. Zhu et al, Nature Neuroscience, May 2020 Targeted by Wave ASOs Variant-selective targeting could address multiple potential drivers of toxicity Neuro C9orf72
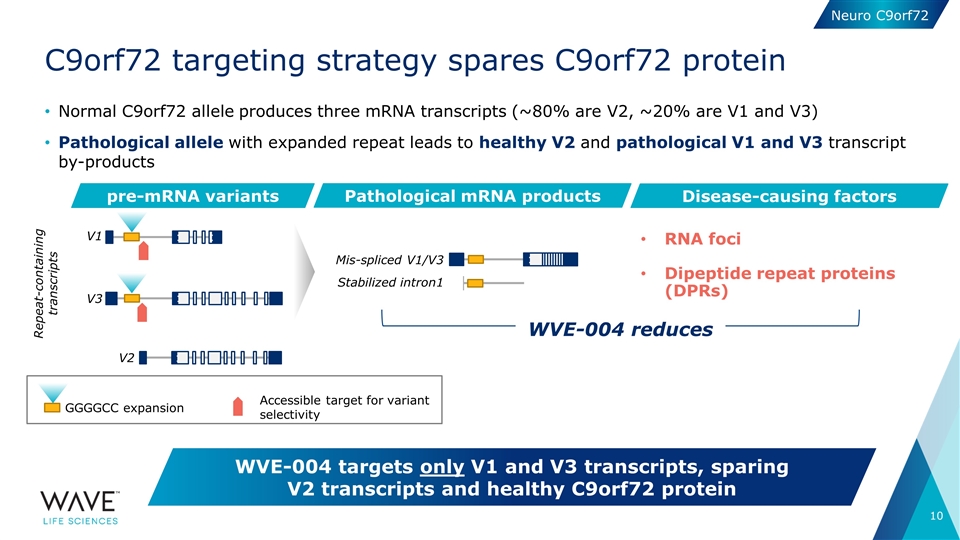
Normal C9orf72 allele produces three mRNA transcripts (~80% are V2, ~20% are V1 and V3) Pathological allele with expanded repeat leads to healthy V2 and pathological V1 and V3 transcript by-products C9orf72 targeting strategy spares C9orf72 protein WVE-004 targets only V1 and V3 transcripts, sparing V2 transcripts and healthy C9orf72 protein pre-mRNA variants Pathological mRNA products V1 V2 Mis-spliced V1/V3 Stabilized intron1 V3 Disease-causing factors RNA foci Dipeptide repeat proteins (DPRs) GGGGCC expansion Accessible target for variant selectivity WVE-004 reduces Repeat-containing transcripts Neuro C9orf72
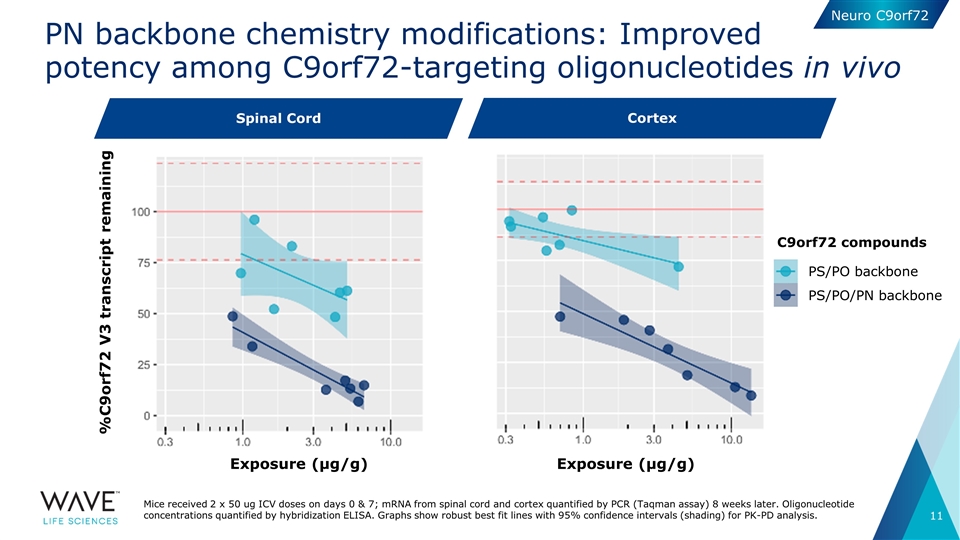
PN backbone chemistry modifications: Improved potency among C9orf72-targeting oligonucleotides in vivo Exposure (µg/g) Exposure (µg/g) C9orf72 compounds Spinal cord Cortex PS/PO backbone PS/PO/PN backbone %C9orf72 V3 transcript remaining Mice received 2 x 50 ug ICV doses on days 0 & 7; mRNA from spinal cord and cortex quantified by PCR (Taqman assay) 8 weeks later. Oligonucleotide concentrations quantified by hybridization ELISA. Graphs show robust best fit lines with 95% confidence intervals (shading) for PK-PD analysis. Spinal Cord Neuro C9orf72
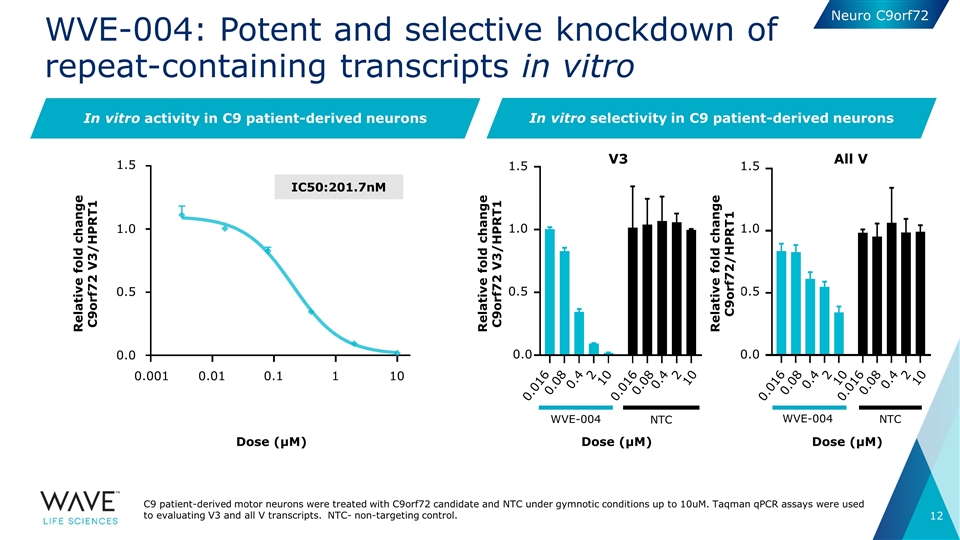
WVE-004: Potent and selective knockdown of repeat-containing transcripts in vitro V3 Dose (μM) All V WVE-004 NTC Dose (μM) In vitro activity in C9 patient-derived neurons WVE-004 NTC Dose (μM) IC50:201.7nM In vitro selectivity in C9 patient-derived neurons C9 patient-derived motor neurons were treated with C9orf72 candidate and NTC under gymnotic conditions up to 10uM. Taqman qPCR assays were used to evaluating V3 and all V transcripts. NTC- non-targeting control. Relative fold change C9orf72 V3/HPRT1 1.5 1.0 0.5 0.0 0.001 0.01 0.1 1 10 Relative fold change C9orf72 V3/HPRT1 0.016 0.08 0.4 2 10 0.016 0.08 0.4 2 10 0.016 0.08 0.4 2 10 0.016 0.08 0.4 2 10 1.5 1.0 0.5 0.0 1.5 1.0 0.5 0.0 Relative fold change C9orf72/HPRT1 Neuro C9orf72
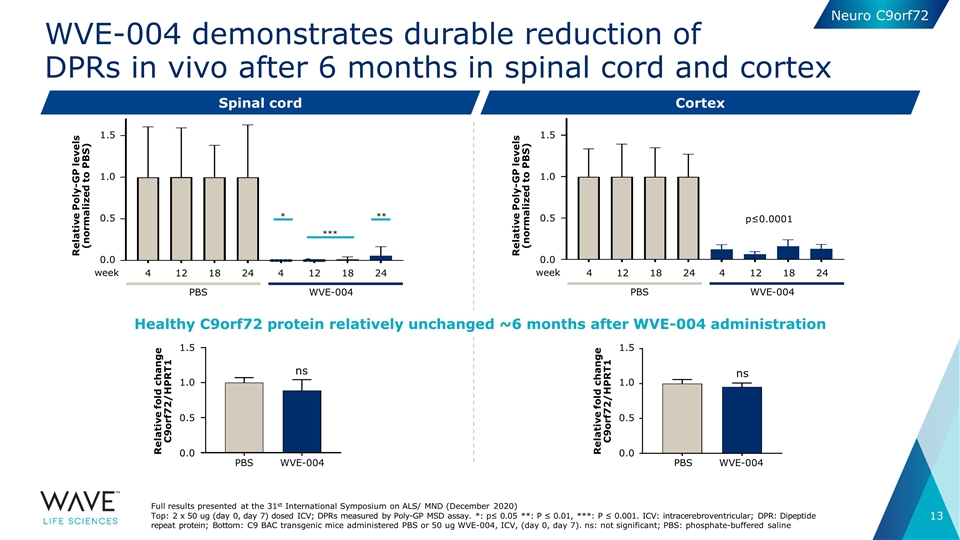
WVE-004 demonstrates durable reduction of DPRs in vivo after 6 months in spinal cord and cortex Spinal cord Cortex Full results presented at the 31st International Symposium on ALS/ MND (December 2020) Top: 2 x 50 ug (day 0, day 7) dosed ICV; DPRs measured by Poly-GP MSD assay. *: p≤ 0.05 **: P ≤ 0.01, ***: P ≤ 0.001. ICV: intracerebroventricular; DPR: Dipeptide repeat protein; Bottom: C9 BAC transgenic mice administered PBS or 50 ug WVE-004, ICV, (day 0, day 7). ns: not significant; PBS: phosphate-buffered saline * *** ** 4 12 18 12 18 24 4 24 WVE-004 PBS week 1.5 0.5 0.0 1.0 Relative Poly-GP levels (normalized to PBS) p≤0.0001 4 12 18 12 18 24 4 24 WVE-004 PBS week 1.5 0.5 0.0 1.0 Relative Poly-GP levels (normalized to PBS) ns Relative fold change C9orf72/HPRT1 1.5 0.5 0.0 1.0 WVE-004 PBS ns Relative fold change C9orf72/HPRT1 1.5 0.5 0.0 1.0 WVE-004 PBS Healthy C9orf72 protein relatively unchanged ~6 months after WVE-004 administration Neuro C9orf72
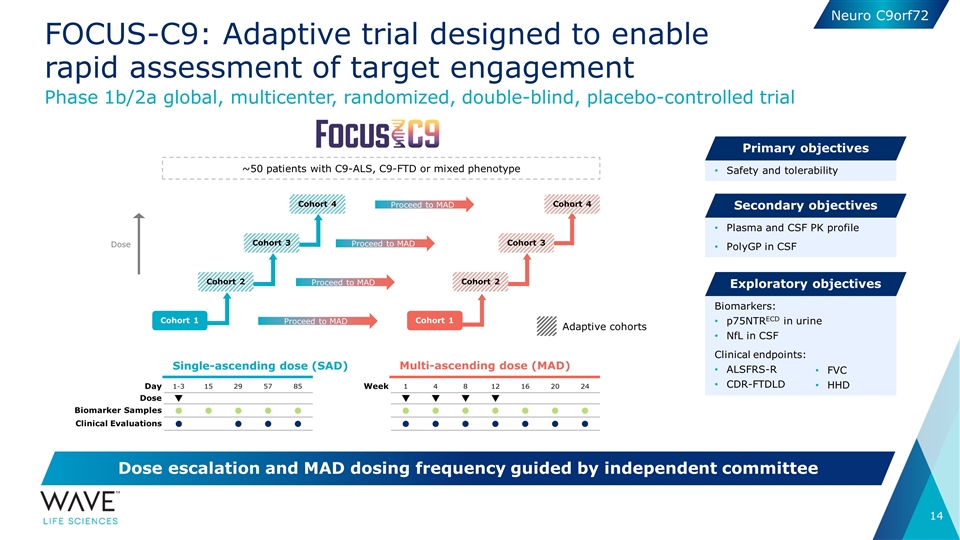
Single-ascending dose (SAD) Multi-ascending dose (MAD) Phase 1b/2a global, multicenter, randomized, double-blind, placebo-controlled trial Dose escalation and MAD dosing frequency guided by independent committee Safety and tolerability Primary objectives Plasma and CSF PK profile PolyGP in CSF Secondary objectives Biomarkers: p75NTRECD in urine NfL in CSF Clinical endpoints: ALSFRS-R CDR-FTDLD Exploratory objectives Day 1-3 15 29 57 85 Dose q Biomarker Samples l l l l l Clinical Evaluations l l l l Week 1 4 8 12 16 20 24 q q q q l l l l l l l l l l l l l l FOCUS-C9: Adaptive trial designed to enable rapid assessment of target engagement ~50 patients with C9-ALS, C9-FTD or mixed phenotype Cohort 1 Proceed to MAD Proceed to MAD Proceed to MAD Cohort 4 Proceed to MAD FVC HHD Adaptive cohorts Cohort 2 Cohort 3 Cohort 1 Cohort 2 Cohort 3 Cohort 4 Dose Neuro C9orf72

WVE-003 Huntington’s Disease
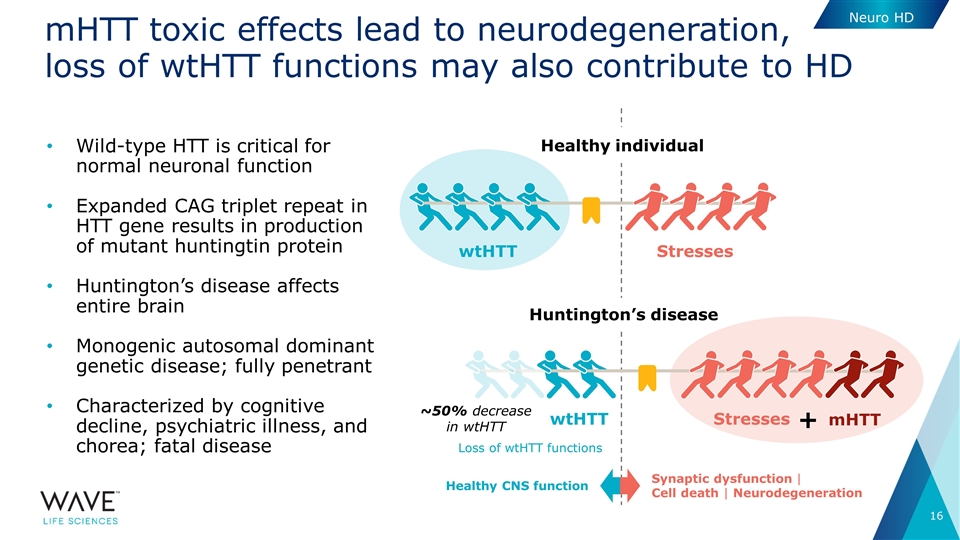
Healthy individual Huntington’s disease mHTT toxic effects lead to neurodegeneration, loss of wtHTT functions may also contribute to HD Wild-type HTT is critical for normal neuronal function Expanded CAG triplet repeat in HTT gene results in production of mutant huntingtin protein Huntington’s disease affects entire brain Monogenic autosomal dominant genetic disease; fully penetrant Characterized by cognitive decline, psychiatric illness, and chorea; fatal disease Stresses wtHTT Stresses wtHTT mHTT + ~50% decrease in wtHTT Healthy CNS function Synaptic dysfunction | Cell death | Neurodegeneration Loss of wtHTT functions Neuro HD
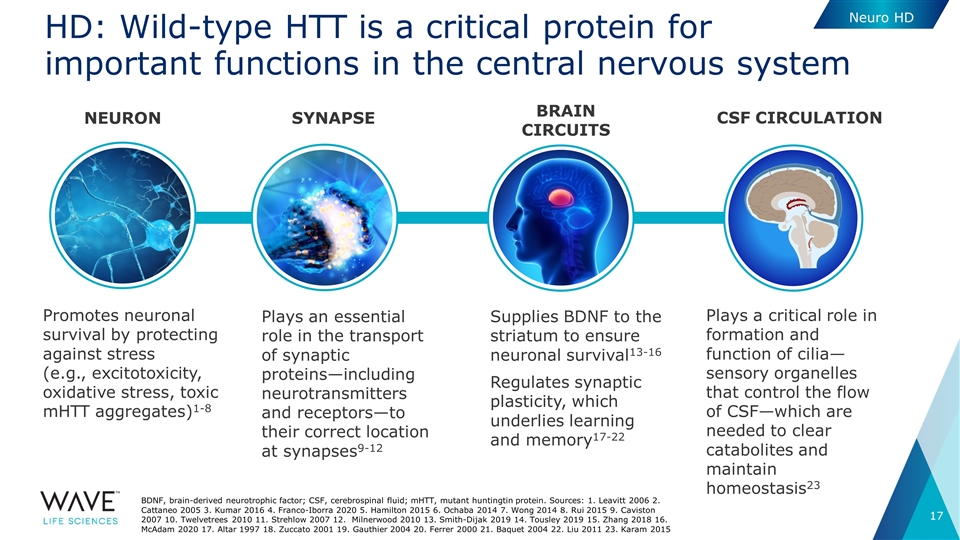
Plays an essential role in the transport of synaptic proteins—including neurotransmitters and receptors—to their correct location at synapses9-12 Promotes neuronal survival by protecting against stress (e.g., excitotoxicity, oxidative stress, toxic mHTT aggregates)1-8 BRAIN CIRCUITS SYNAPSE NEURON CSF circulation Supplies BDNF to the striatum to ensure neuronal survival13-16 Regulates synaptic plasticity, which underlies learning and memory17-22 Plays a critical role in formation and function of cilia—sensory organelles that control the flow of CSF—which are needed to clear catabolites and maintain homeostasis23 HD: Wild-type HTT is a critical protein for important functions in the central nervous system BDNF, brain-derived neurotrophic factor; CSF, cerebrospinal fluid; mHTT, mutant huntingtin protein. Sources: 1. Leavitt 2006 2. Cattaneo 2005 3. Kumar 2016 4. Franco-Iborra 2020 5. Hamilton 2015 6. Ochaba 2014 7. Wong 2014 8. Rui 2015 9. Caviston 2007 10. Twelvetrees 2010 11. Strehlow 2007 12. Milnerwood 2010 13. Smith-Dijak 2019 14. Tousley 2019 15. Zhang 2018 16. McAdam 2020 17. Altar 1997 18. Zuccato 2001 19. Gauthier 2004 20. Ferrer 2000 21. Baquet 2004 22. Liu 2011 23. Karam 2015 Neuro HD
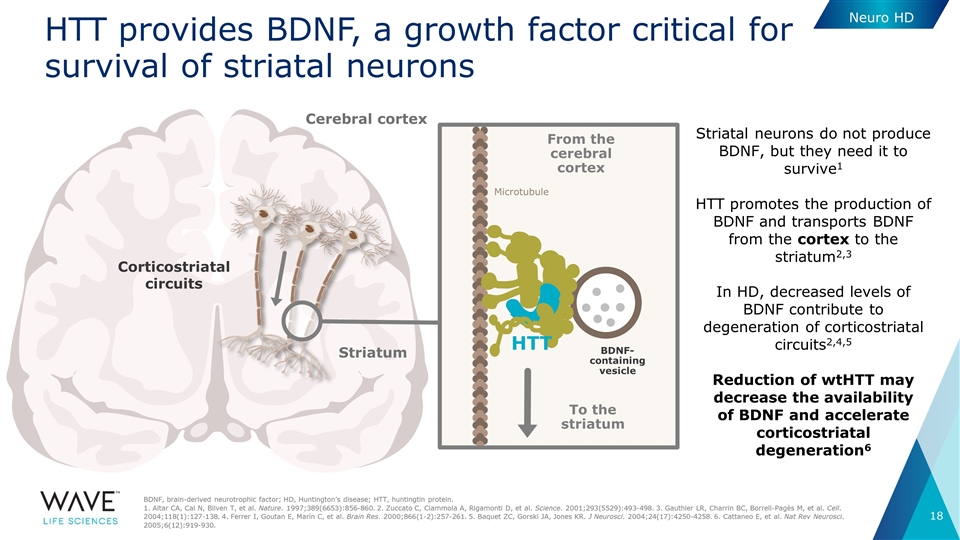
Cerebral cortex Striatum BDNF- containing vesicle To the striatum Microtubule HTT HTT provides BDNF, a growth factor critical for survival of striatal neurons BDNF, brain-derived neurotrophic factor; HD, Huntington’s disease; HTT, huntingtin protein. 1. Altar CA, Cai N, Bliven T, et al. Nature. 1997;389(6653):856-860. 2. Zuccato C, Ciammola A, Rigamonti D, et al. Science. 2001;293(5529):493-498. 3. Gauthier LR, Charrin BC, Borrell-Pagès M, et al. Cell. 2004;118(1):127-138. 4. Ferrer I, Goutan E, Marín C, et al. Brain Res. 2000;866(1-2):257-261. 5. Baquet ZC, Gorski JA, Jones KR. J Neurosci. 2004;24(17):4250-4258. 6. Cattaneo E, et al. Nat Rev Neurosci. 2005;6(12):919-930. From the cerebral cortex Striatal neurons do not produce BDNF, but they need it to survive1 HTT promotes the production of BDNF and transports BDNF from the cortex to the striatum2,3 In HD, decreased levels of BDNF contribute to degeneration of corticostriatal circuits2,4,5 Reduction of wtHTT may decrease the availability of BDNF and accelerate corticostriatal degeneration6 Corticostriatal circuits Neuro HD
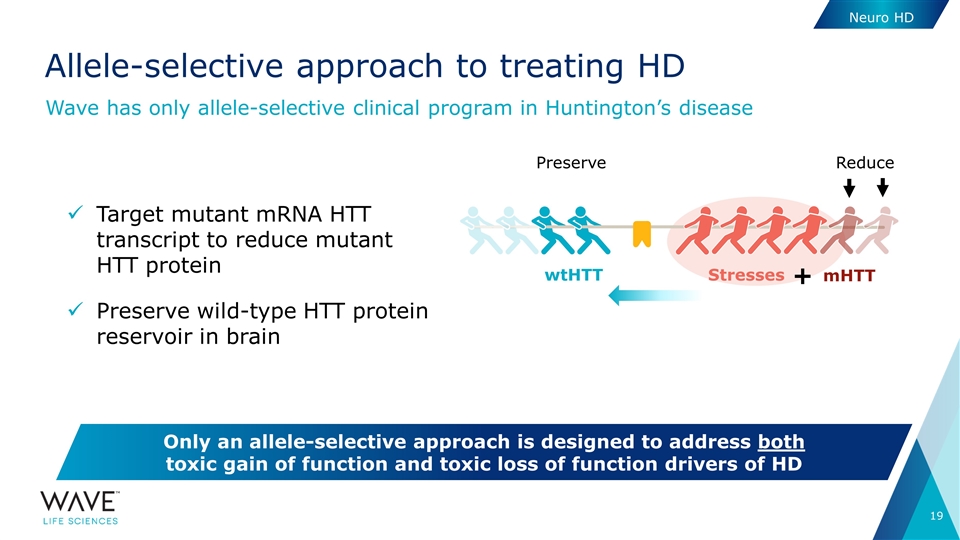
Target mutant mRNA HTT transcript to reduce mutant HTT protein Preserve wild-type HTT protein reservoir in brain Allele-selective approach to treating HD Wave has only allele-selective clinical program in Huntington’s disease Only an allele-selective approach is designed to address both toxic gain of function and toxic loss of function drivers of HD Stresses wtHTT mHTT + Reduce Preserve Neuro HD
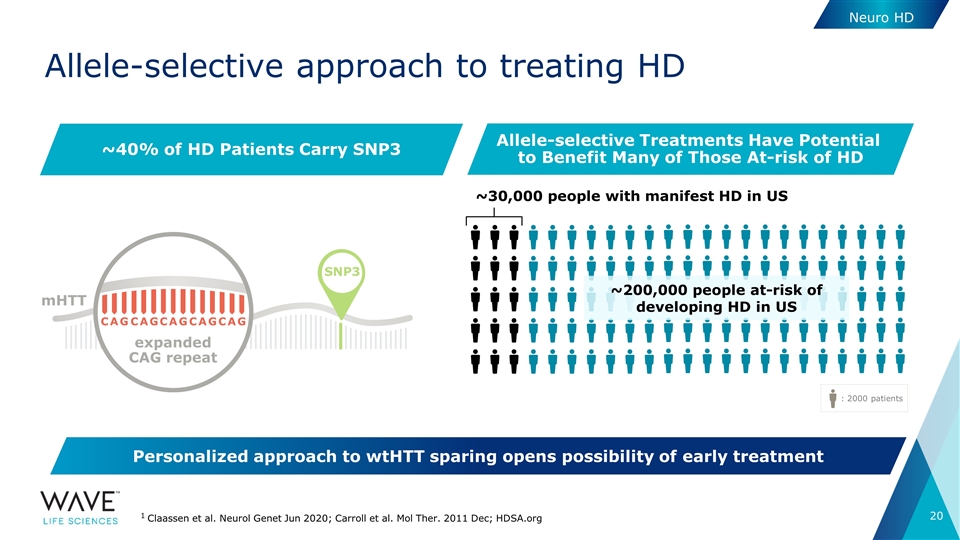
Allele-selective approach to treating HD Neuro HD 1 Claassen et al. Neurol Genet Jun 2020; Carroll et al. Mol Ther. 2011 Dec; HDSA.org : 2000 patients ~30,000 people with manifest HD in US ~40% of HD Patients Carry SNP3 Allele-selective Treatments Have Potential to Benefit Many of Those At-risk of HD ~200,000 people at-risk of developing HD in US SNP3 C A G C A G C A G C A G C A G expanded CAG repeat mHTT Personalized approach to wtHTT sparing opens possibility of early treatment
Nature publication contributes to weight of evidence on importance of wild-type huntingtin Source: Poplawski et al., Nature, April 2019 Htt: Huntingtin protein Conditional knock-out of Htt in 4-month old mice (post-neuronal development) Results suggest that: Htt plays a central role in the regenerating transcriptome (potentially influencing genes such as NFKB, STAT3, BDNF) Htt is essential for regeneration Indeed, conditional gene deletion showed that Htt is required for neuronal repair. Throughout life, neuronal maintenance and repair are essential to support adequate cellular functioning Neuro HD
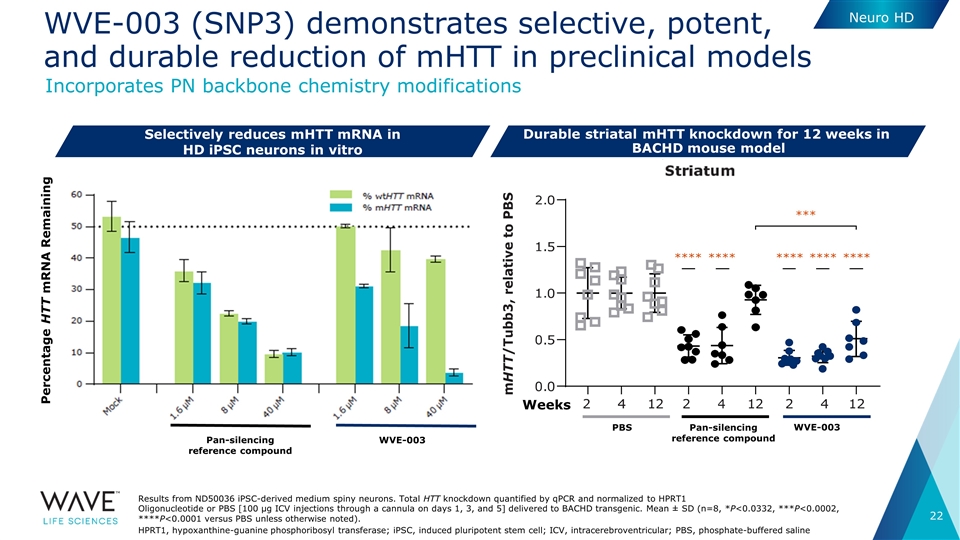
WVE-003 (SNP3) demonstrates selective, potent, and durable reduction of mHTT in preclinical models Selectively reduces mHTT mRNA in HD iPSC neurons in vitro Results from ND50036 iPSC-derived medium spiny neurons. Total HTT knockdown quantified by qPCR and normalized to HPRT1 Oligonucleotide or PBS [100 μg ICV injections through a cannula on days 1, 3, and 5] delivered to BACHD transgenic. Mean ± SD (n=8, *P<0.0332, ***P<0.0002, ****P<0.0001 versus PBS unless otherwise noted). HPRT1, hypoxanthine-guanine phosphoribosyl transferase; iPSC, induced pluripotent stem cell; ICV, intracerebroventricular; PBS, phosphate-buffered saline Similar results in cortex Pan-silencing reference compound WVE-003 PBS Weeks *** **** **** **** **** **** Pan-silencing reference compound WVE-003 Percentage HTT mRNA Remaining Durable striatal mHTT knockdown for 12 weeks in BACHD mouse model Neuro HD Incorporates PN backbone chemistry modifications
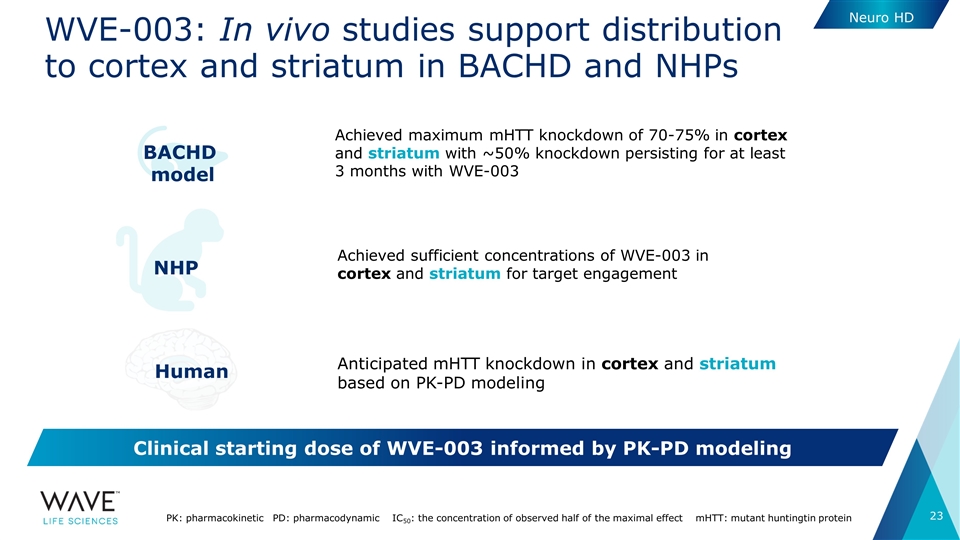
WVE-003: In vivo studies support distribution to cortex and striatum in BACHD and NHPs PK: pharmacokinetic PD: pharmacodynamic IC50: the concentration of observed half of the maximal effect mHTT: mutant huntingtin protein Achieved sufficient concentrations of WVE-003 in cortex and striatum for target engagement NHP Anticipated mHTT knockdown in cortex and striatum based on PK-PD modeling Human BACHD model Achieved maximum mHTT knockdown of 70-75% in cortex and striatum with ~50% knockdown persisting for at least 3 months with WVE-003 Clinical starting dose of WVE-003 informed by PK-PD modeling Neuro HD
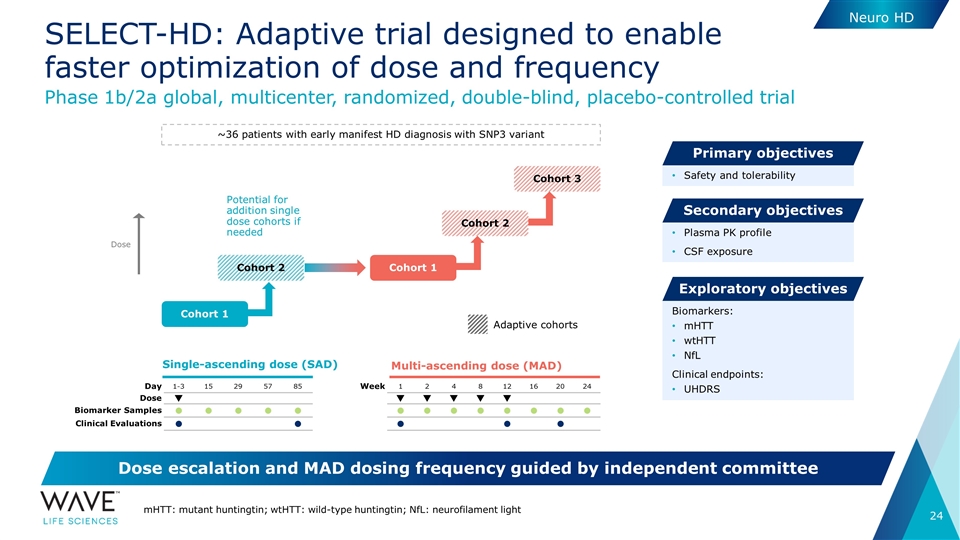
Single-ascending dose (SAD) Multi-ascending dose (MAD) Day 1-3 15 29 57 85 Dose q Biomarker Samples l l l l l Clinical Evaluations l l Week 1 2 4 8 12 16 20 24 q q q q q l l l l l l l l l l l SELECT-HD: Adaptive trial designed to enable faster optimization of dose and frequency Adaptive cohorts Phase 1b/2a global, multicenter, randomized, double-blind, placebo-controlled trial Safety and tolerability Primary objectives Plasma PK profile CSF exposure Secondary objectives Biomarkers: mHTT wtHTT NfL Clinical endpoints: UHDRS Exploratory objectives Dose escalation and MAD dosing frequency guided by independent committee mHTT: mutant huntingtin; wtHTT: wild-type huntingtin; NfL: neurofilament light ~36 patients with early manifest HD diagnosis with SNP3 variant Cohort 1 Cohort 2 Cohort 1 Cohort 2 Cohort 3 Potential for addition single dose cohorts if needed Dose Neuro HD
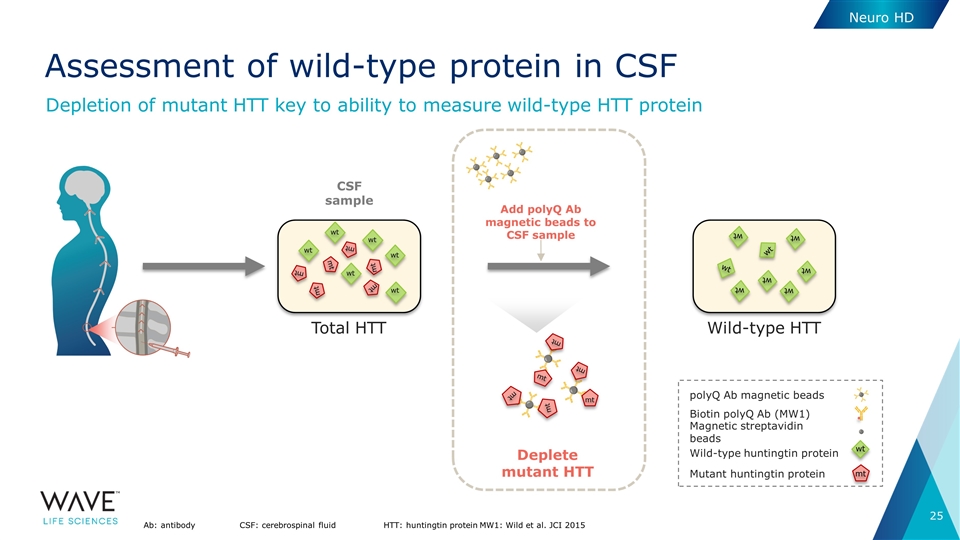
Neuro HD Assessment of wild-type protein in CSF Ab: antibodyCSF: cerebrospinal fluidHTT: huntingtin proteinMW1: Wild et al. JCI 2015 CSF sample Total HTT mt wt wt wt mt mt mt mt mt wt wt wt Wild-type HTT wt wt wt wt wt wt wt wt Deplete mutant HTT Add polyQ Ab magnetic beads to CSF sample mt mt mt mt mt mt polyQ Ab magnetic beads Biotin polyQ Ab (MW1) Magnetic streptavidin beads Wild-type huntingtin protein Mutant huntingtin protein wt mt Depletion of mutant HTT key to ability to measure wild-type HTT protein

WVE-N531 Duchenne muscular dystrophy
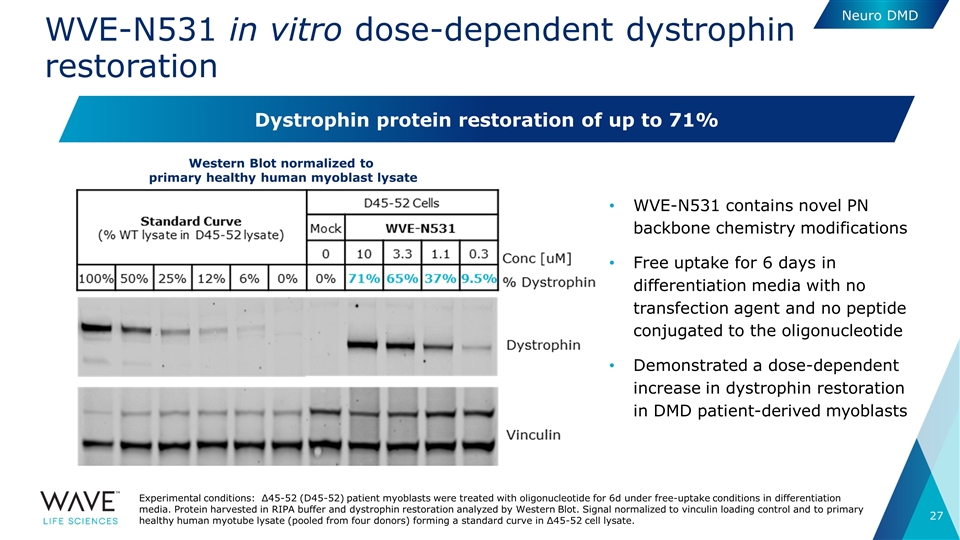
WVE-N531 in vitro dose-dependent dystrophin restoration WVE-N531 contains novel PN backbone chemistry modifications Free uptake for 6 days in differentiation media with no transfection agent and no peptide conjugated to the oligonucleotide Demonstrated a dose-dependent increase in dystrophin restoration in DMD patient-derived myoblasts Experimental conditions: Δ45-52 (D45-52) patient myoblasts were treated with oligonucleotide for 6d under free-uptake conditions in differentiation media. Protein harvested in RIPA buffer and dystrophin restoration analyzed by Western Blot. Signal normalized to vinculin loading control and to primary healthy human myotube lysate (pooled from four donors) forming a standard curve in Δ45-52 cell lysate. Western Blot normalized to primary healthy human myoblast lysate Dystrophin protein restoration of up to 71% Neuro DMD
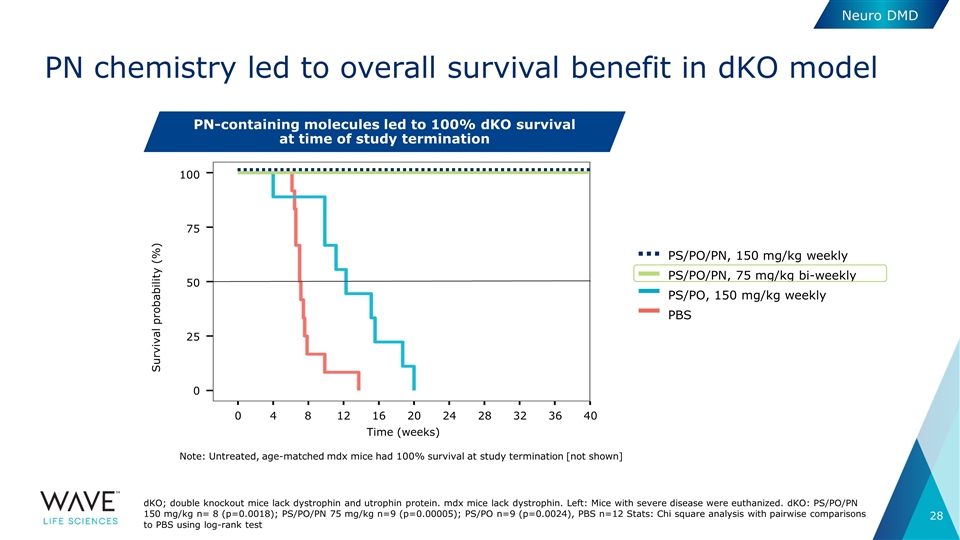
PN chemistry led to overall survival benefit in dKO model dKO; double knockout mice lack dystrophin and utrophin protein. mdx mice lack dystrophin. Left: Mice with severe disease were euthanized. dKO: PS/PO/PN 150 mg/kg n= 8 (p=0.0018); PS/PO/PN 75 mg/kg n=9 (p=0.00005); PS/PO n=9 (p=0.0024), PBS n=12 Stats: Chi square analysis with pairwise comparisons to PBS using log-rank test PN-containing molecules led to 100% dKO survival at time of study termination PS/PO/PN, 75 mg/kg bi-weekly PBS PS/PO, 150 mg/kg weekly PS/PO/PN, 150 mg/kg weekly 100 75 50 25 0 Survival probability (%) 0 4 8 12 16 20 24 28 32 36 40 Time (weeks) Note: Untreated, age-matched mdx mice had 100% survival at study termination [not shown] Neuro DMD

Clinical trial of WVE-N531 to initiate in 2021 Unmet need in DMD remains high CTA submitted in March 2021 to initiate clinical development Clinical trial powered to evaluate change in dystrophin production, and will assess drug concentration in muscle, and initial safety Open-label study; targeting every-other-week administration in up to 15 boys with DMD Potential to apply PN chemistry to other exons if successful Dosing in clinical trial expected to initiate in 2021 Neuro DMD

Wave’s discovery and drug development platform
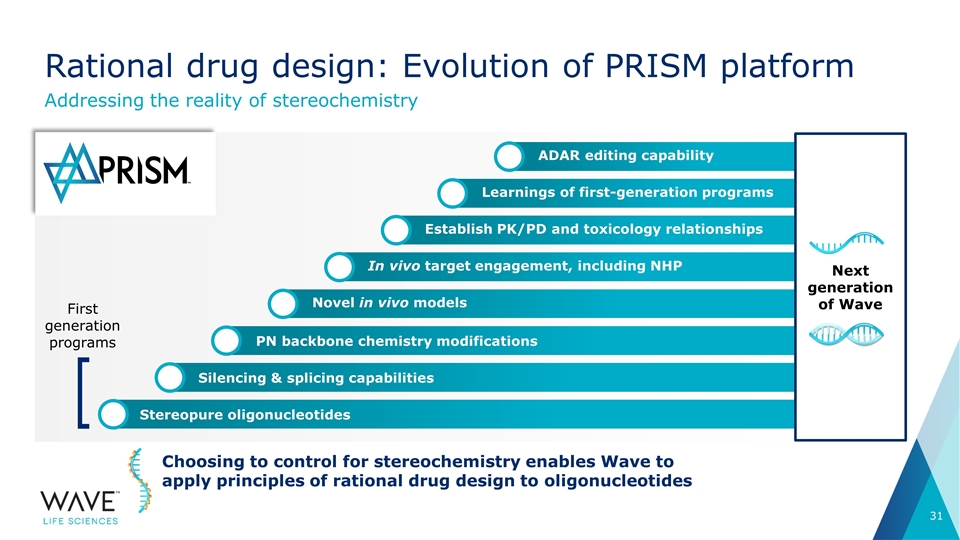
Silencing & splicing capabilities PN backbone chemistry modifications In vivo target engagement, including NHP ADAR editing capability Learnings of first-generation programs Establish PK/PD and toxicology relationships Novel in vivo models Stereopure oligonucleotides Rational drug design: Evolution of PRISM platform Addressing the reality of stereochemistry Choosing to control for stereochemistry enables Wave to apply principles of rational drug design to oligonucleotides First generation programs Next generation of Wave
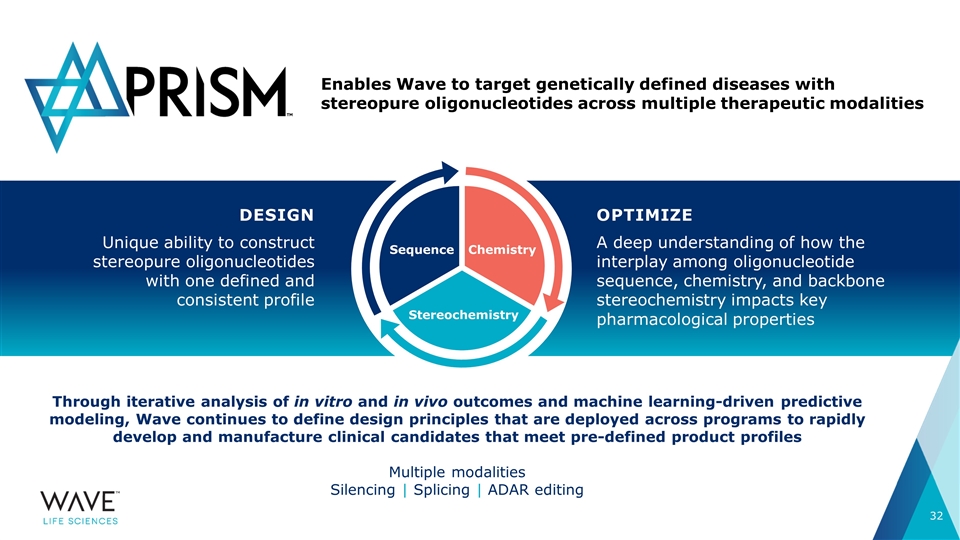
Through iterative analysis of in vitro and in vivo outcomes and machine learning-driven predictive modeling, Wave continues to define design principles that are deployed across programs to rapidly develop and manufacture clinical candidates that meet pre-defined product profiles Multiple modalities Silencing | Splicing | ADAR editing DESIGN Unique ability to construct stereopure oligonucleotides with one defined and consistent profile Enables Wave to target genetically defined diseases with stereopure oligonucleotides across multiple therapeutic modalities OPTIMIZE A deep understanding of how the interplay among oligonucleotide sequence, chemistry, and backbone stereochemistry impacts key pharmacological properties SEQUENCE STEREOCHEMISTRY CHEMISTRY Sequence Stereochemistry Chemistry
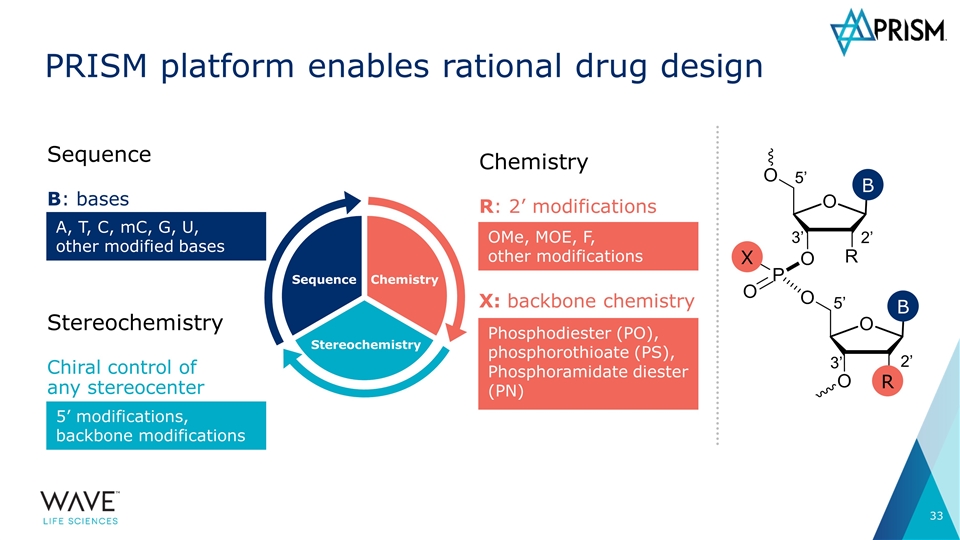
Sequence Stereochemistry Chemistry PRISM platform enables rational drug design Chemistry R: 2’ modifications OMe, MOE, F, other modifications 5’ 2’ 3’ 5’ 3’ 2’ R X B B X: backbone chemistry Phosphodiester (PO), phosphorothioate (PS), Phosphoramidate diester (PN) Sequence B: bases A, T, C, mC, G, U, other modified bases Stereochemistry Chiral control of any stereocenter 5’ modifications, backbone modifications
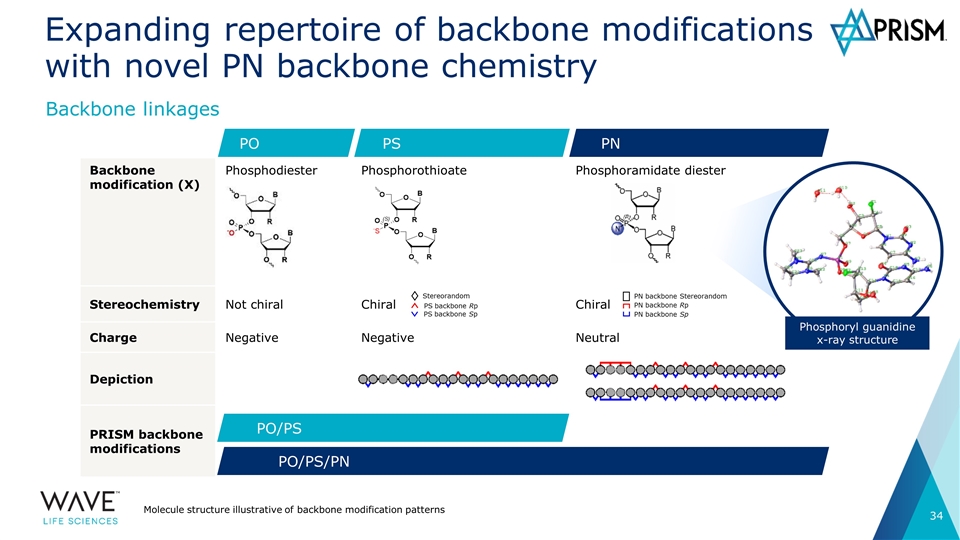
Backbone modification (X) Phosphodiester Phosphorothioate Phosphoramidate diester Stereochemistry Not chiral Chiral Chiral Charge Negative Negative Neutral Depiction PRISM backbone modifications Expanding repertoire of backbone modifications with novel PN backbone chemistry Molecule structure illustrative of backbone modification patterns Backbone linkages PS PO PN PO/PS PO/PS/PN Phosphoryl guanidine x-ray structure Stereorandom PS backbone Rp PS backbone Sp PN backbone Sp PN backbone Rp PN backbone Stereorandom
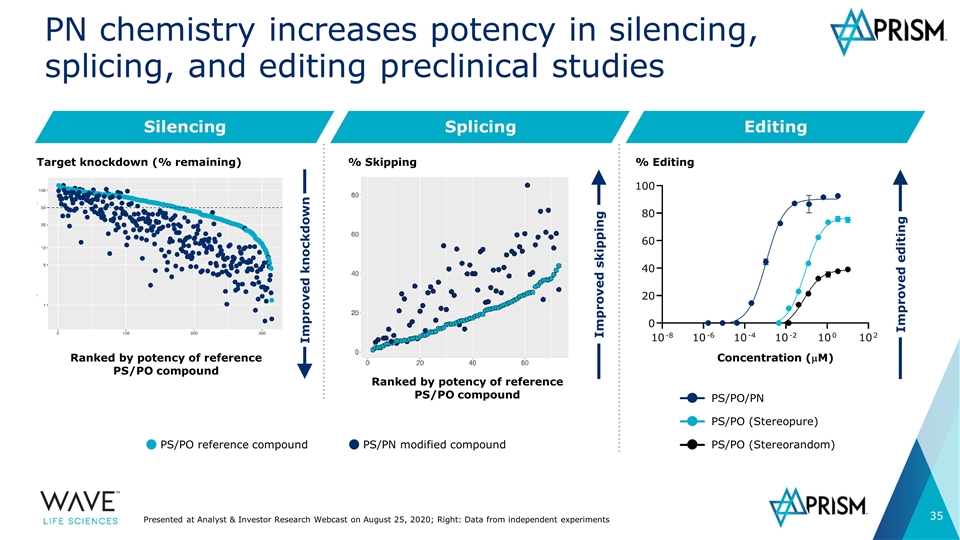
Silencing PN chemistry increases potency in silencing, splicing, and editing preclinical studies Improved knockdown Splicing Editing Improved skipping Ranked by potency of reference PS/PO compound Ranked by potency of reference PS/PO compound Improved editing PS/PO/PN PS/PO (Stereopure) PS/PO (Stereorandom) Concentration (mM) % Editing PS/PO reference compound PS/PN modified compound % Skipping Target knockdown (% remaining) Presented at Analyst & Investor Research Webcast on August 25, 2020; Right: Data from independent experiments
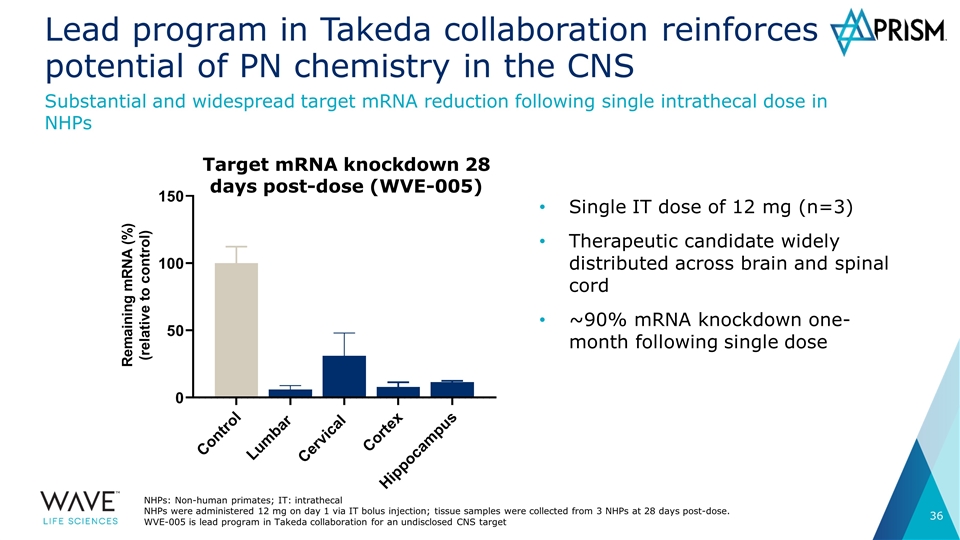
Lead program in Takeda collaboration reinforces potential of PN chemistry in the CNS Single IT dose of 12 mg (n=3) Therapeutic candidate widely distributed across brain and spinal cord ~90% mRNA knockdown one-month following single dose Substantial and widespread target mRNA reduction following single intrathecal dose in NHPs NHPs: Non-human primates; IT: intrathecal NHPs were administered 12 mg on day 1 via IT bolus injection; tissue samples were collected from 3 NHPs at 28 days post-dose. WVE-005 is lead program in Takeda collaboration for an undisclosed CNS target Target mRNA knockdown 28 days post-dose (WVE-005)
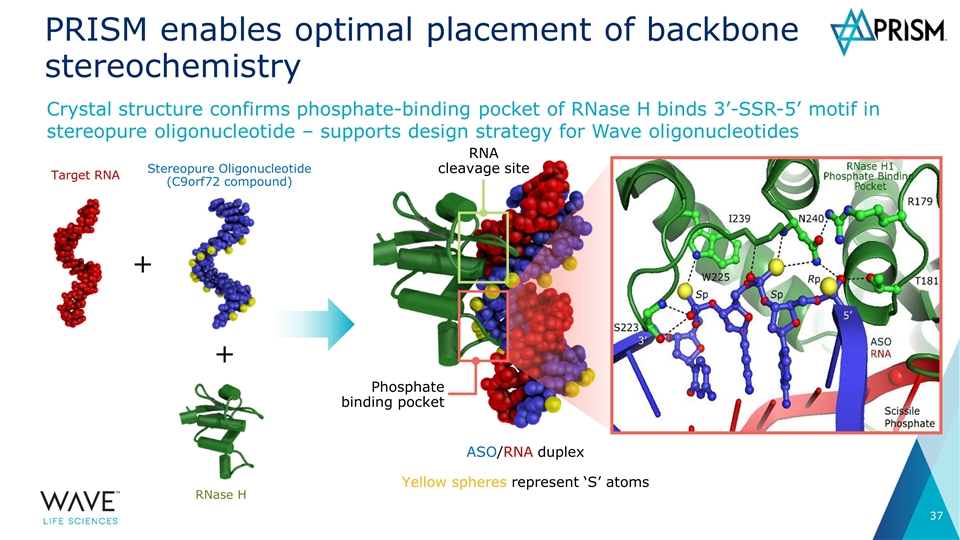
PRISM enables optimal placement of backbone stereochemistry Crystal structure confirms phosphate-binding pocket of RNase H binds 3’-SSR-5’ motif in stereopure oligonucleotide – supports design strategy for Wave oligonucleotides ASO/RNA duplex Yellow spheres represent ‘S’ atoms Phosphate binding pocket RNA cleavage site Target RNA Stereopure Oligonucleotide (C9orf72 compound) RNase H + +
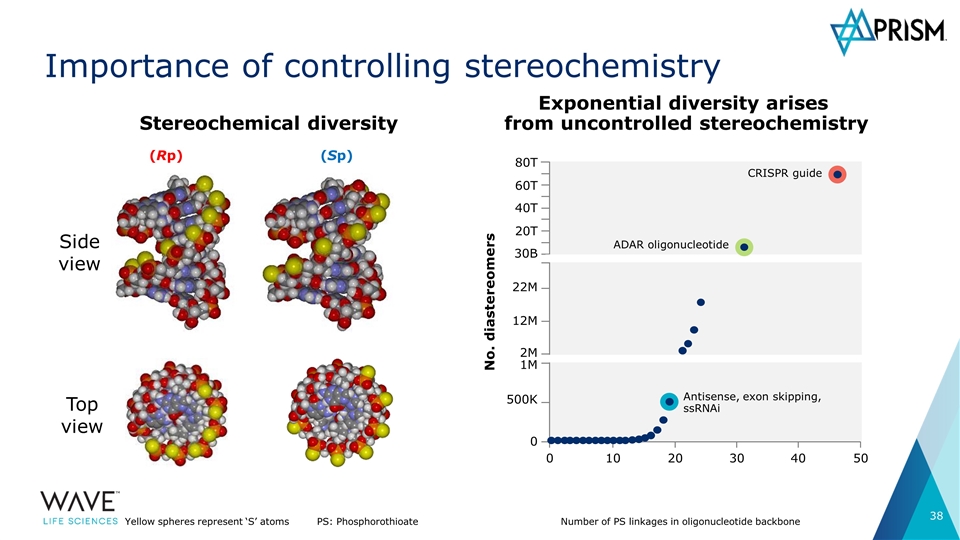
Importance of controlling stereochemistry (Rp) (Sp) Top view Side view Yellow spheres represent ‘S’ atomsPS: Phosphorothioate Number of PS linkages in oligonucleotide backbone No. diastereomers 80T 60T 40T 20T 30B 22M 12M 2M 1M 500K 0 0 10 20 30 40 50 Antisense, exon skipping, ssRNAi ADAR oligonucleotide CRISPR guide Stereochemical diversity Exponential diversity arises from uncontrolled stereochemistry

ADAR editing Platform capability and Alpha-1 antitrypsin deficiency
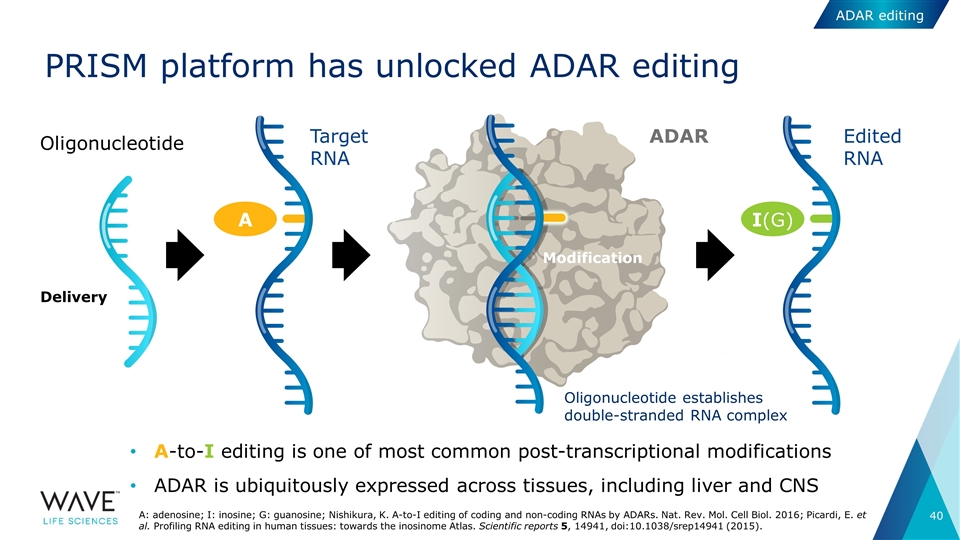
PRISM platform has unlocked ADAR editing A-to-I editing is one of most common post-transcriptional modifications ADAR is ubiquitously expressed across tissues, including liver and CNS ADAR Target RNA I(G) A Edited RNA Oligonucleotide establishes double-stranded RNA complex Oligonucleotide Modification Delivery A: adenosine; I: inosine; G: guanosine; Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016; Picardi, E. et al. Profiling RNA editing in human tissues: towards the inosinome Atlas. Scientific reports 5, 14941, doi:10.1038/srep14941 (2015). ADAR editing
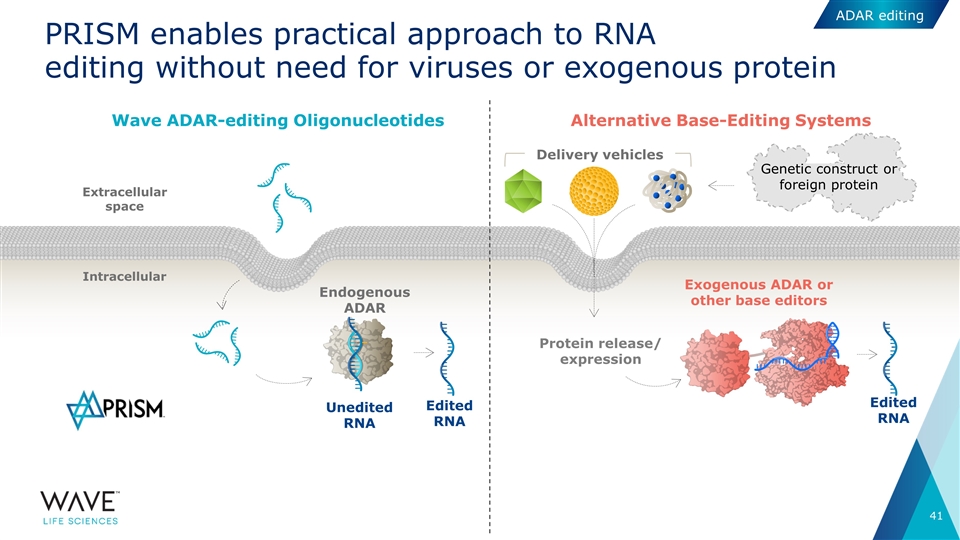
PRISM enables practical approach to RNA editing without need for viruses or exogenous protein Intracellular Extracellular space Endogenous ADAR Unedited RNA Wave ADAR-editing Oligonucleotides Exogenous ADAR or other base editors Edited RNA Protein release/ expression Delivery vehicles Alternative Base-Editing Systems Edited RNA Genetic construct or foreign protein ADAR editing
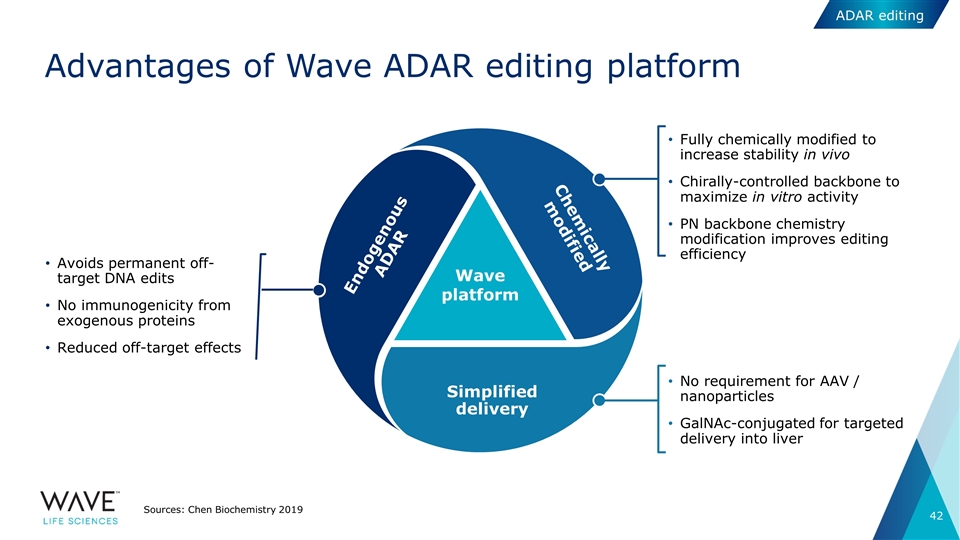
Wave platform Fully chemically modified to increase stability in vivo Chirally-controlled backbone to maximize in vitro activity PN backbone chemistry modification improves editing efficiency No requirement for AAV / nanoparticles GalNAc-conjugated for targeted delivery into liver Avoids permanent off-target DNA edits No immunogenicity from exogenous proteins Reduced off-target effects Advantages of Wave ADAR editing platform Sources: Chen Biochemistry 2019 Chemically modified Simplified delivery Endogenous ADAR ADAR editing
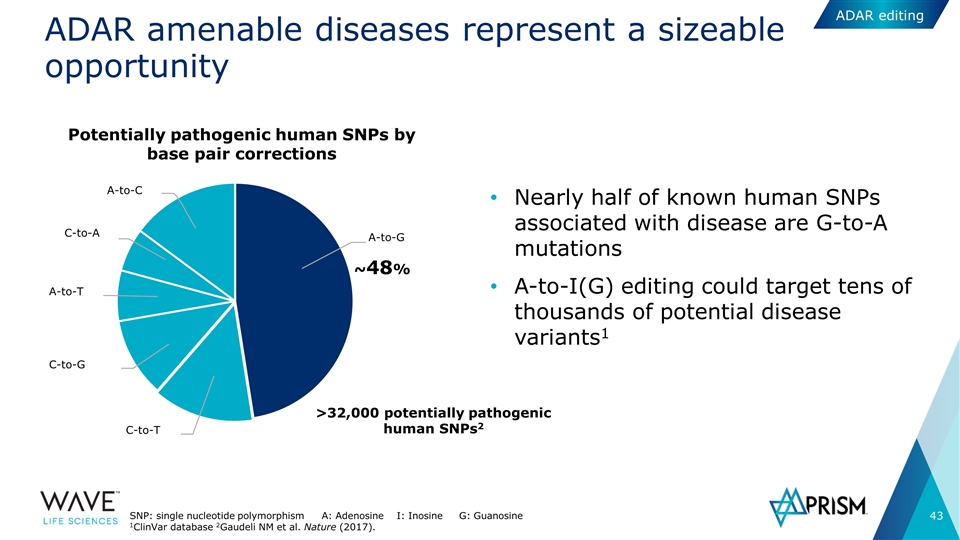
ADAR amenable diseases represent a sizeable opportunity Nearly half of known human SNPs associated with disease are G-to-A mutations A-to-I(G) editing could target tens of thousands of potential disease variants1 ~48% C-to-T C-to-G A-to-T C-to-A A-to-C A-to-G Potentially pathogenic human SNPs by base pair corrections >32,000 potentially pathogenic human SNPs2 SNP: single nucleotide polymorphism A: Adenosine I: Inosine G: Guanosine 1ClinVar database 2Gaudeli NM et al. Nature (2017). ADAR editing
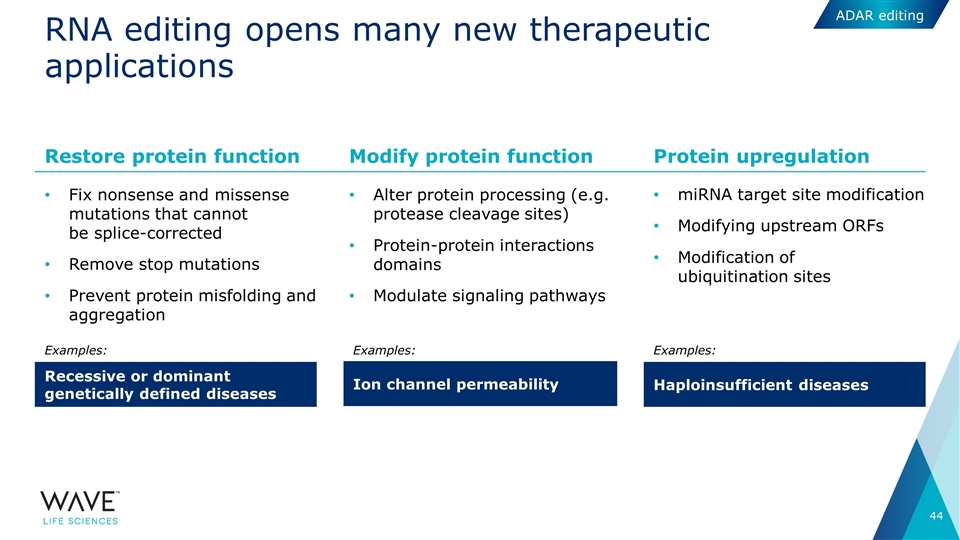
RNA editing opens many new therapeutic applications Fix nonsense and missense mutations that cannot be splice-corrected Remove stop mutations Prevent protein misfolding and aggregation Alter protein processing (e.g. protease cleavage sites) Protein-protein interactions domains Modulate signaling pathways miRNA target site modification Modifying upstream ORFs Modification of ubiquitination sites Restore protein function Recessive or dominant genetically defined diseases Modify protein function Ion channel permeability Protein upregulation Haploinsufficient diseases Examples: Examples: Examples: ADAR editing
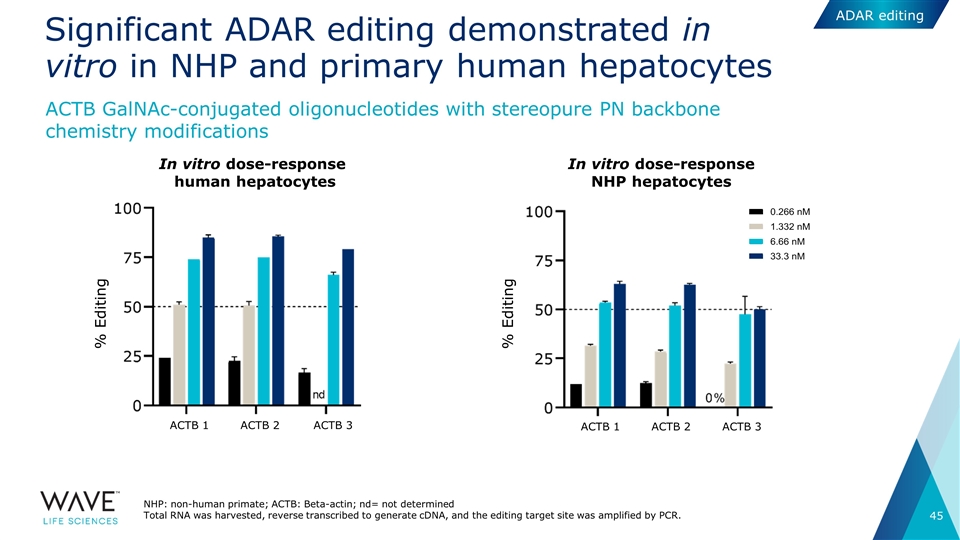
Significant ADAR editing demonstrated in vitro in NHP and primary human hepatocytes NHP: non-human primate; ACTB: Beta-actin; nd= not determined Total RNA was harvested, reverse transcribed to generate cDNA, and the editing target site was amplified by PCR. In vitro dose-response human hepatocytes In vitro dose-response NHP hepatocytes % Editing % Editing ACTB 1 ACTB 2 ACTB 3 ACTB 1 ACTB 2 ACTB 3 ACTB GalNAc-conjugated oligonucleotides with stereopure PN backbone chemistry modifications ADAR editing
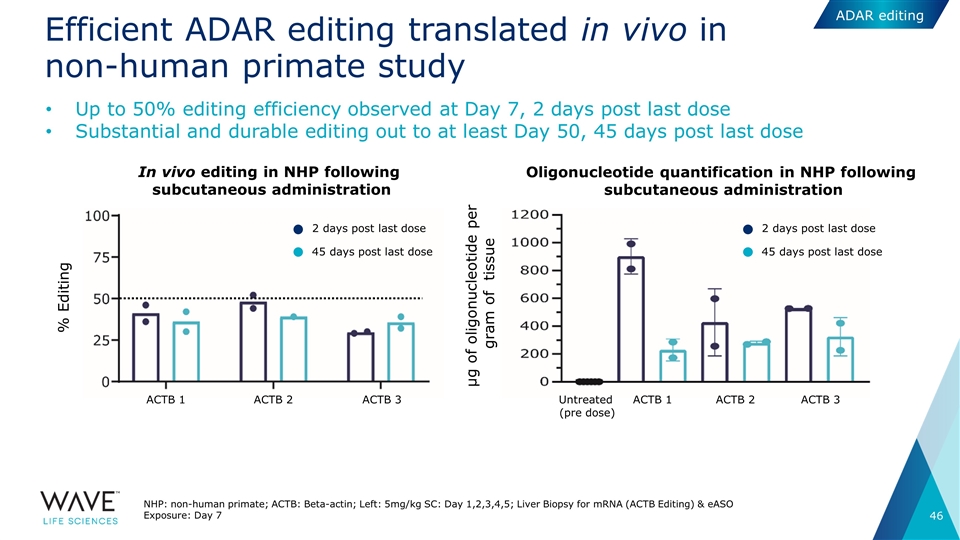
Efficient ADAR editing translated in vivo in non-human primate study NHP: non-human primate; ACTB: Beta-actin; Left: 5mg/kg SC: Day 1,2,3,4,5; Liver Biopsy for mRNA (ACTB Editing) & eASO Exposure: Day 7 Up to 50% editing efficiency observed at Day 7, 2 days post last dose Substantial and durable editing out to at least Day 50, 45 days post last dose In vivo editing in NHP following subcutaneous administration Oligonucleotide quantification in NHP following subcutaneous administration 2 days post last dose 45 days post last dose 2 days post last dose 45 days post last dose ACTB 1 ACTB 2 ACTB 3 ACTB 1 ACTB 2 ACTB 3 Untreated (pre dose) % Editing µg of oligonucleotide per gram of tissue ADAR editing
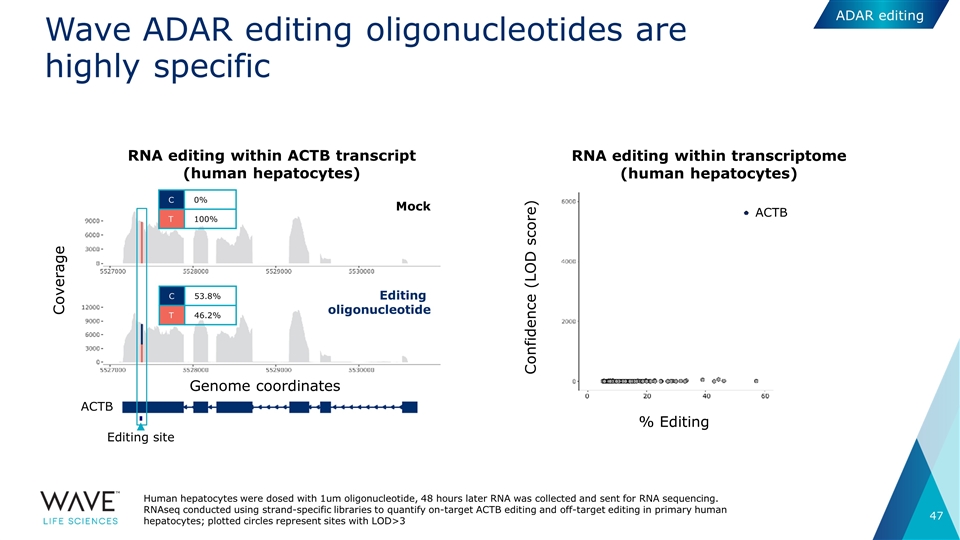
Editing site RNA editing within ACTB transcript (human hepatocytes) RNA editing within transcriptome (human hepatocytes) Wave ADAR editing oligonucleotides are highly specific Coverage Genome coordinates ACTB C 0% T 100% C 53.8% T 46.2% ACTB Confidence (LOD score) % Editing Mock Editing oligonucleotide Human hepatocytes were dosed with 1um oligonucleotide, 48 hours later RNA was collected and sent for RNA sequencing. RNAseq conducted using strand-specific libraries to quantify on-target ACTB editing and off-target editing in primary human hepatocytes; plotted circles represent sites with LOD>3 ADAR editing
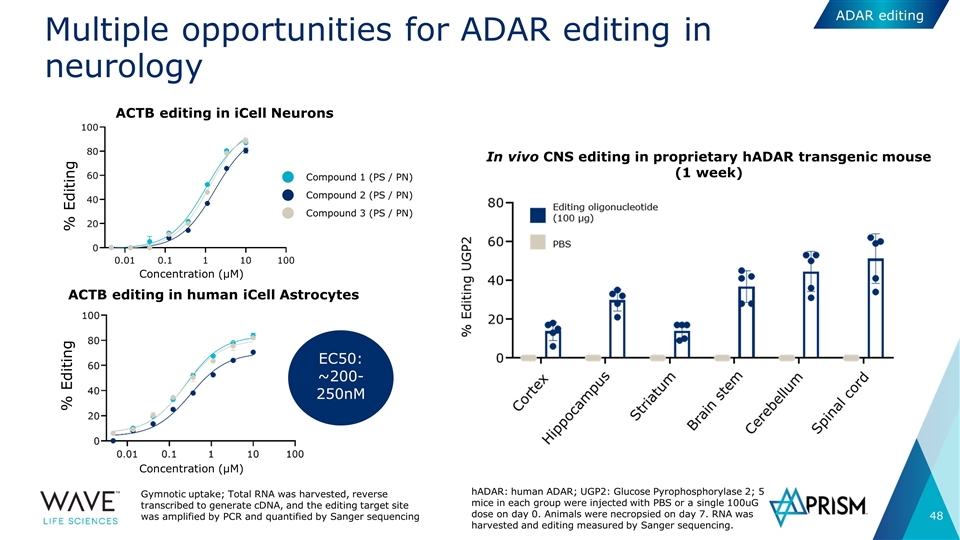
Multiple opportunities for ADAR editing in neurology ACTB editing in iCell Neurons ACTB editing in human iCell Astrocytes Concentration (µM) % Editing % Editing Compound 2 (PS / PN) Compound 1 (PS / PN) Compound 3 (PS / PN) EC50: ~200-250nM Gymnotic uptake; Total RNA was harvested, reverse transcribed to generate cDNA, and the editing target site was amplified by PCR and quantified by Sanger sequencing Concentration (µM) ADAR editing hADAR: human ADAR; UGP2: Glucose Pyrophosphorylase 2; 5 mice in each group were injected with PBS or a single 100uG dose on day 0. Animals were necropsied on day 7. RNA was harvested and editing measured by Sanger sequencing. In vivo CNS editing in proprietary hADAR transgenic mouse (1 week)
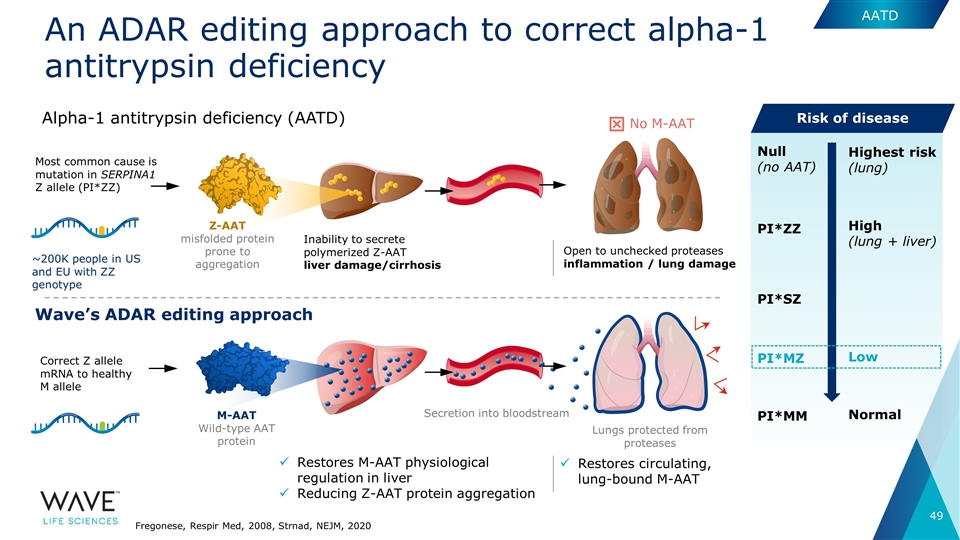
An ADAR editing approach to correct alpha-1 antitrypsin deficiency Correct Z allele mRNA to healthy M allele Most common cause is mutation in SERPINA1 Z allele (PI*ZZ) Z-AAT misfolded protein prone to aggregation Restores circulating, lung-bound M-AAT M-AAT Wild-type AAT protein Secretion into bloodstream Lungs protected from proteases Restores M-AAT physiological regulation in liver Reducing Z-AAT protein aggregation Wave’s ADAR editing approach Alpha-1 antitrypsin deficiency (AATD) AATD Inability to secrete polymerized Z-AAT liver damage/cirrhosis Open to unchecked proteases inflammation / lung damage No M-AAT PI*MM Normal PI*MZ Low PI*SZ PI*ZZ High (lung + liver) Null (no AAT) Highest risk (lung) Fregonese, Respir Med, 2008, Strnad, NEJM, 2020 Risk of disease ~200K people in US and EU with ZZ genotype
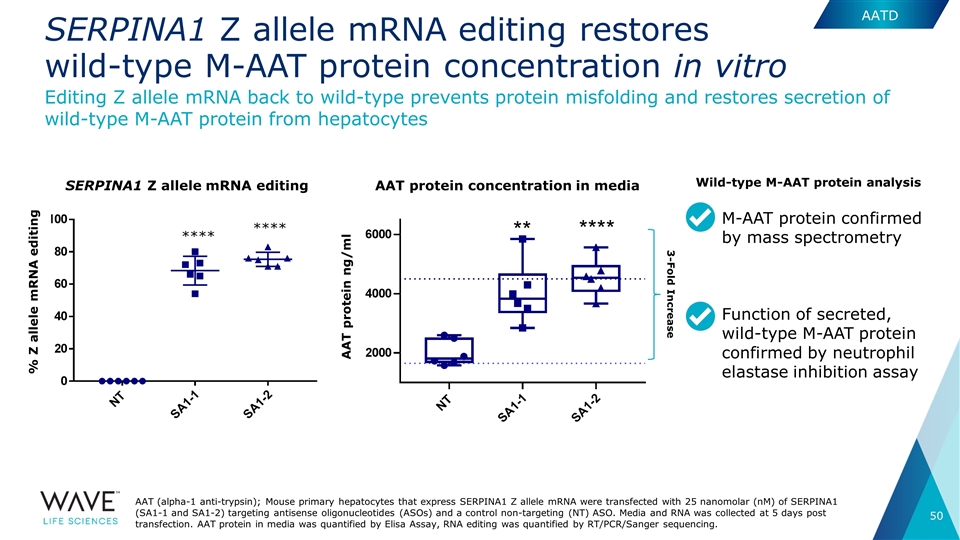
3-Fold Increase SERPINA1 Z allele mRNA editing restores wild-type M-AAT protein concentration in vitro AAT (alpha-1 anti-trypsin); Mouse primary hepatocytes that express SERPINA1 Z allele mRNA were transfected with 25 nanomolar (nM) of SERPINA1 (SA1-1 and SA1-2) targeting antisense oligonucleotides (ASOs) and a control non-targeting (NT) ASO. Media and RNA was collected at 5 days post transfection. AAT protein in media was quantified by Elisa Assay, RNA editing was quantified by RT/PCR/Sanger sequencing. Editing Z allele mRNA back to wild-type prevents protein misfolding and restores secretion of wild-type M-AAT protein from hepatocytes AAT protein concentration in media SERPINA1 Z allele mRNA editing % Z allele mRNA editing AAT protein ng/ml M-AAT protein confirmed by mass spectrometry Function of secreted, wild-type M-AAT protein confirmed by neutrophil elastase inhibition assay Wild-type M-AAT protein analysis AATD **** ****
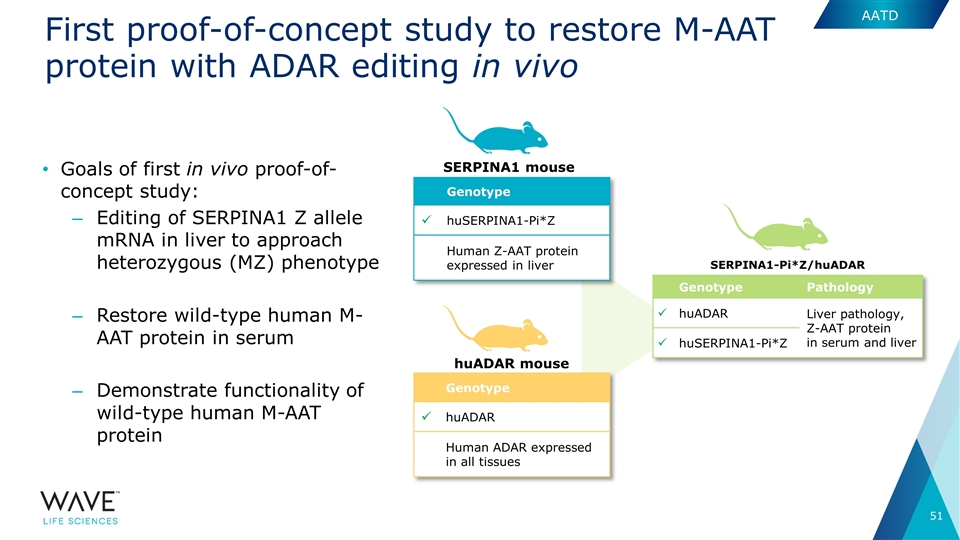
First proof-of-concept study to restore M-AAT protein with ADAR editing in vivo SERPINA1-Pi*Z/huADAR Genotype Pathology ü huADAR Liver pathology, Z-AAT protein in serum and liver ü huSERPINA1-Pi*Z Goals of first in vivo proof-of-concept study: Editing of SERPINA1 Z allele mRNA in liver to approach heterozygous (MZ) phenotype Restore wild-type human M-AAT protein in serum Demonstrate functionality of wild-type human M-AAT protein Genotype ü huADAR Human ADAR expressed in all tissues huADAR mouse SERPINA1 mouse Genotype ü huSERPINA1-Pi*Z Human Z-AAT protein expressed in liver AATD
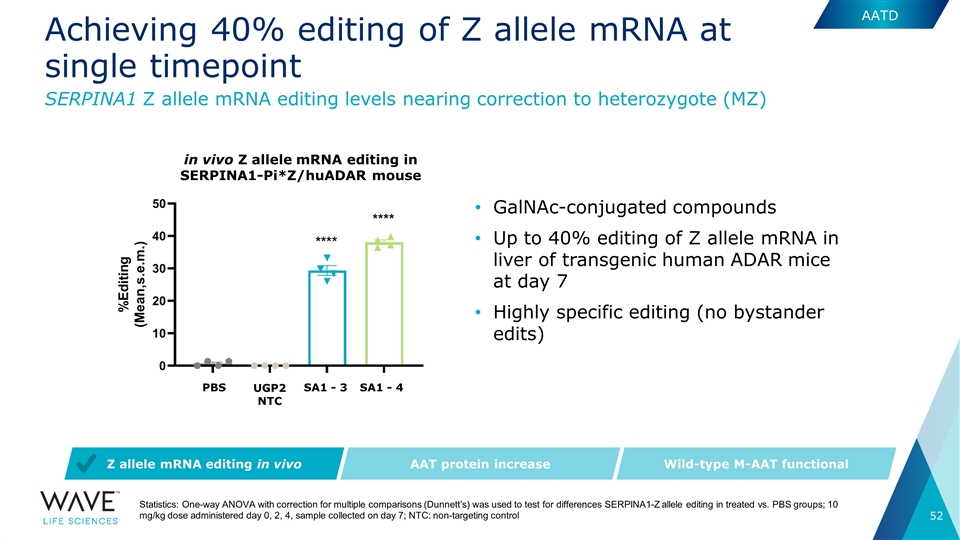
Achieving 40% editing of Z allele mRNA at single timepoint SA1 - 3 SA1 - 4 SERPINA1 Z allele mRNA editing levels nearing correction to heterozygote (MZ) AAT protein increase Wild-type M-AAT functional Z allele mRNA editing in vivo GalNAc-conjugated compounds Up to 40% editing of Z allele mRNA in liver of transgenic human ADAR mice at day 7 Highly specific editing (no bystander edits) Statistics: One-way ANOVA with correction for multiple comparisons (Dunnett’s) was used to test for differences SERPINA1-Z allele editing in treated vs. PBS groups; 10 mg/kg dose administered day 0, 2, 4, sample collected on day 7; NTC: non-targeting control in vivo Z allele mRNA editing in SERPINA1-Pi*Z/huADAR mouse UGP2 NTC PBS AATD
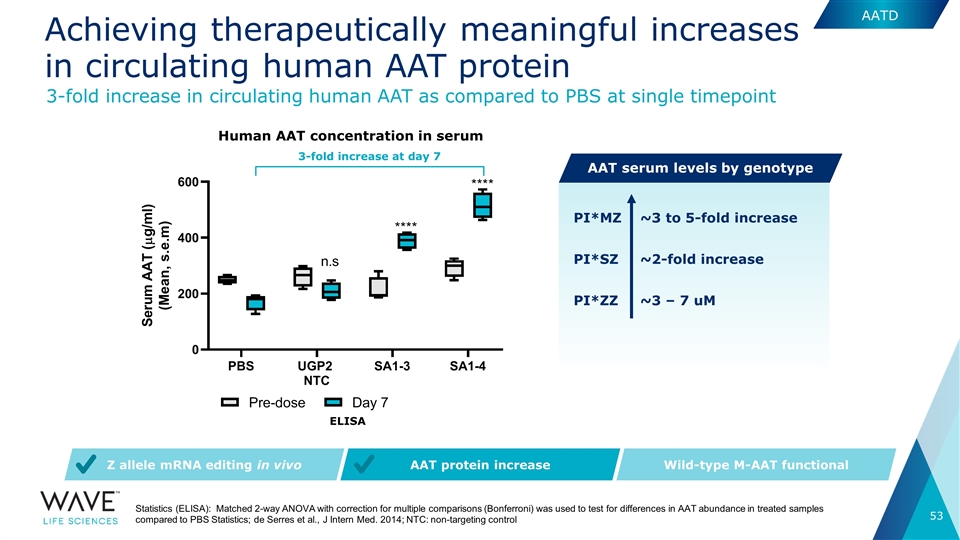
Statistics (ELISA): Matched 2-way ANOVA with correction for multiple comparisons (Bonferroni) was used to test for differences in AAT abundance in treated samples compared to PBS Statistics; de Serres et al., J Intern Med. 2014; NTC: non-targeting control AAT protein increase Wild-type M-AAT functional Z allele mRNA editing in vivo PI*SZ ~2-fold increase ~3 to 5-fold increase PI*MZ 3-fold increase at day 7 Human AAT concentration in serum ELISA PI*ZZ ~3 – 7 uM 3-fold increase in circulating human AAT as compared to PBS at single timepoint AATD AAT serum levels by genotype Achieving therapeutically meaningful increases in circulating human AAT protein
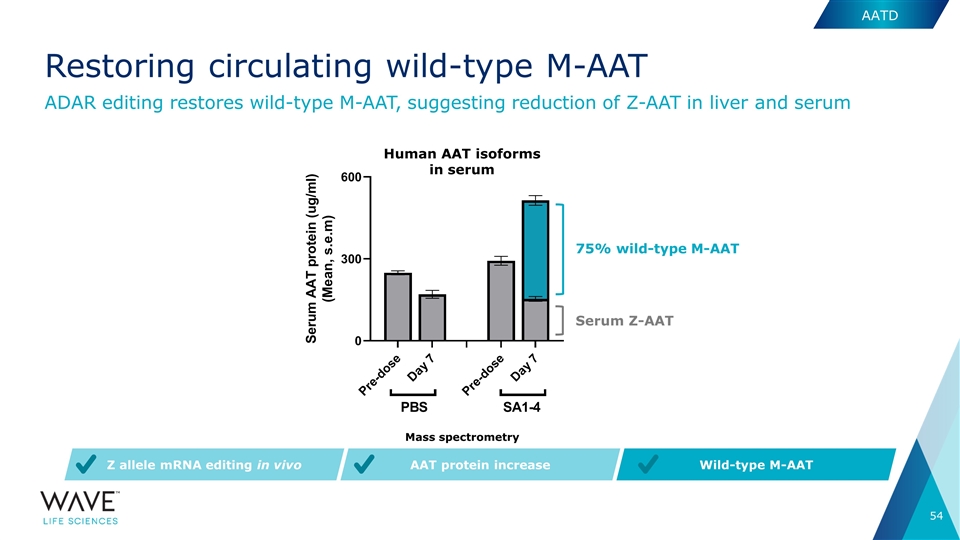
75% wild-type M-AAT Serum Z-AAT AAT protein increase Wild-type M-AAT Z allele mRNA editing in vivo Human AAT isoforms in serum Mass spectrometry ADAR editing restores wild-type M-AAT, suggesting reduction of Z-AAT in liver and serum AATD Restoring circulating wild-type M-AAT
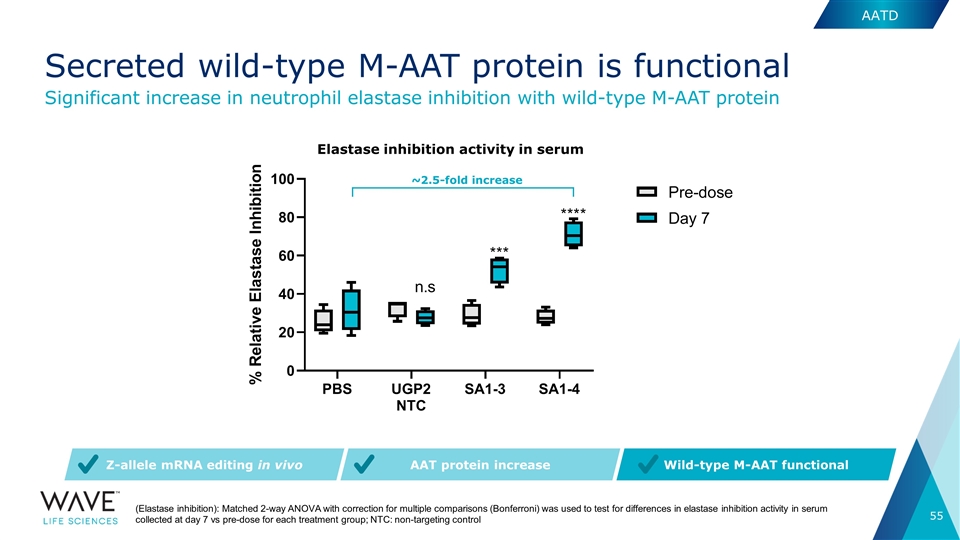
(Elastase inhibition): Matched 2-way ANOVA with correction for multiple comparisons (Bonferroni) was used to test for differences in elastase inhibition activity in serum collected at day 7 vs pre-dose for each treatment group; NTC: non-targeting control AAT protein increase Wild-type M-AAT functional Z-allele mRNA editing in vivo ~2.5-fold increase Elastase inhibition activity in serum Significant increase in neutrophil elastase inhibition with wild-type M-AAT protein AATD Secreted wild-type M-AAT protein is functional
ADAR editing successfully corrects Z allele mRNA in vivo to restore functional M-AAT protein Results support use of human transgenic mouse model to evaluate ADAR editing compounds for additional targets Up to 40% editing of SERPINA1 Z allele mRNA in liver at single timepoint, nearing correction to heterozygotes (MZ) Initial Z allele mRNA editing resulted in therapeutically meaningful increase in circulating functional wild-type M-AAT protein in vivo Restoration of wild-type M-AAT suggests reduction of mutant Z-AAT protein in liver and serum Ongoing studies to assess duration of activity, dose response, PK/PD, reduction in Z-AAT protein aggregates to provide insight into wild-type M-AAT secretion levels over time and changes in liver pathology Additional data on durability and dose response expected in 2H 2021 AATD

Ophthalmology
Stereopure oligonucleotides for inherited retinal diseases (IRDs) Wave ophthalmology opportunity Oligonucleotides can be administered by intravitreal (IVT) injection; targeting twice per year dosing Stereopure oligonucleotides open novel strategies in both dominant and recessive IRDs; potential for potent and durable effect with low immune response Successful targeting of MALAT1 is a surrogate for an ASO mechanism of action Widely expressed in many different cell types Only expressed in the nucleus Intravitreal injection Sources: Daiger S, et al. Clin Genet. 2013;84:132-141. Wong CH, et al. Biostatistics. 2018; DOI: 10.1093/biostatistics/kxx069. Athanasiou D, et al. Prog Retin Eye Res. 2018;62:1–23. Daiger S, et al. Cold Spring Harb Perspect Med. 2015;5:a017129. Verbakel S, et al. Prog Retin Eye Res. 2018:66:157-186.; Short, B.G.; Toxicology Pathology, Jan 2008. Ophthalmology
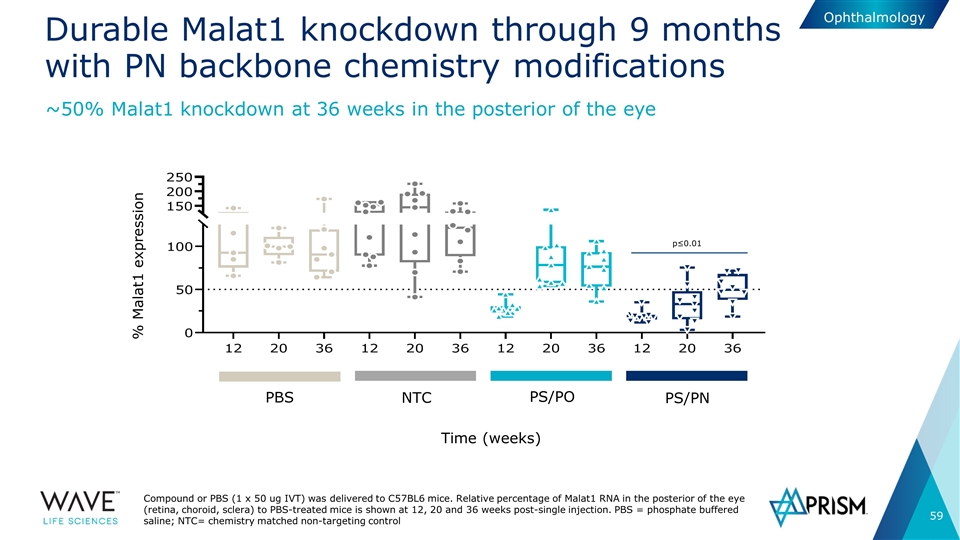
Durable Malat1 knockdown through 9 months with PN backbone chemistry modifications Compound or PBS (1 x 50 ug IVT) was delivered to C57BL6 mice. Relative percentage of Malat1 RNA in the posterior of the eye (retina, choroid, sclera) to PBS-treated mice is shown at 12, 20 and 36 weeks post-single injection. PBS = phosphate buffered saline; NTC= chemistry matched non-targeting control ~50% Malat1 knockdown at 36 weeks in the posterior of the eye PBS NTC PS/PO PS/PN % Malat1 expression Time (weeks) p≤0.01 Ophthalmology

Usher Syndrome Type 2A: a progressive vision loss disorder Autosomal recessive disease characterized by hearing loss at birth and progressive vision loss beginning in adolescence or adulthood Caused by mutations in USH2A gene (72 exons) that disrupt production of usherin protein in retina, leading to degeneration of the photoreceptors No approved disease-modifying therapies ~5,000 addressable patients in US Sources: Boughman et al., 1983. J Chron Dis. 36:595-603; Seyedahmadi et al., 2004. Exp Eye Res. 79:167-173; Liu et al., 2007. Proc Natl Acad Sci USA 104:4413-4418. Oligonucleotides that promote USH2A exon 13 skipping may restore production of functional usherin protein Ophthalmology
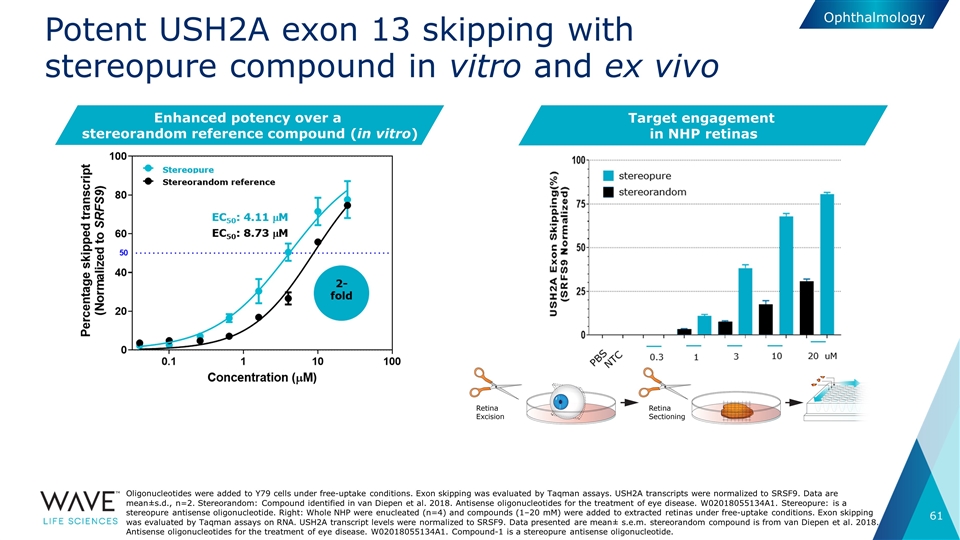
Potent USH2A exon 13 skipping with stereopure compound in vitro and ex vivo Oligonucleotides were added to Y79 cells under free-uptake conditions. Exon skipping was evaluated by Taqman assays. USH2A transcripts were normalized to SRSF9. Data are mean±s.d., n=2. Stereorandom: Compound identified in van Diepen et al. 2018. Antisense oligonucleotides for the treatment of eye disease. W02018055134A1. Stereopure: is a stereopure antisense oligonucleotide. Right: Whole NHP were enucleated (n=4) and compounds (1–20 mM) were added to extracted retinas under free-uptake conditions. Exon skipping was evaluated by Taqman assays on RNA. USH2A transcript levels were normalized to SRSF9. Data presented are mean± s.e.m. stereorandom compound is from van Diepen et al. 2018. Antisense oligonucleotides for the treatment of eye disease. W02018055134A1. Compound-1 is a stereopure antisense oligonucleotide. Enhanced potency over a stereorandom reference compound (in vitro) Ophthalmology Target engagement in NHP retinas
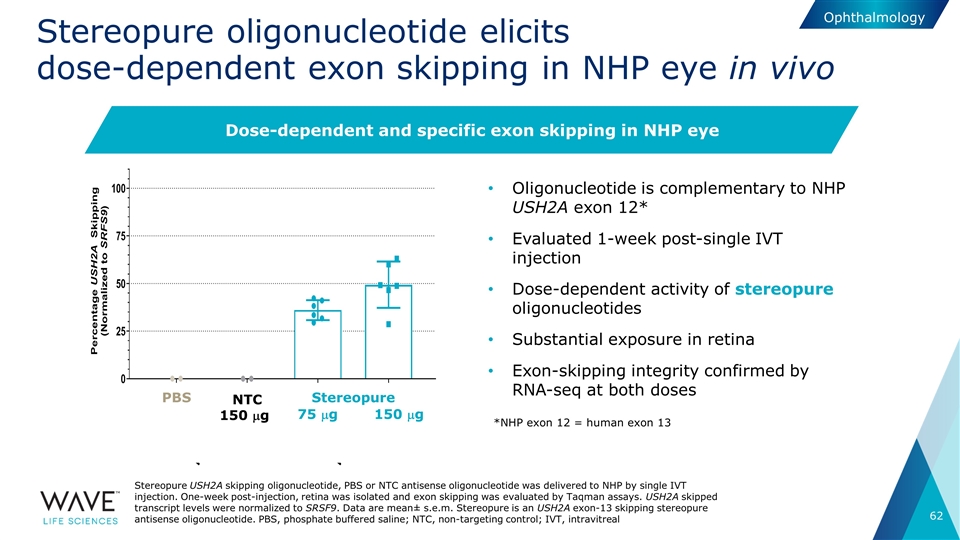
Stereopure oligonucleotide elicits dose-dependent exon skipping in NHP eye in vivo Oligonucleotide is complementary to NHP USH2A exon 12* Evaluated 1-week post-single IVT injection Dose-dependent activity of stereopure oligonucleotides Substantial exposure in retina Exon-skipping integrity confirmed by RNA-seq at both doses NTC 150 mg 75 mg 150 mg PBS Stereopure Stereopure USH2A skipping oligonucleotide, PBS or NTC antisense oligonucleotide was delivered to NHP by single IVT injection. One-week post-injection, retina was isolated and exon skipping was evaluated by Taqman assays. USH2A skipped transcript levels were normalized to SRSF9. Data are mean± s.e.m. Stereopure is an USH2A exon-13 skipping stereopure antisense oligonucleotide. PBS, phosphate buffered saline; NTC, non-targeting control; IVT, intravitreal Dose-dependent and specific exon skipping in NHP eye *NHP exon 12 = human exon 13 Ophthalmology
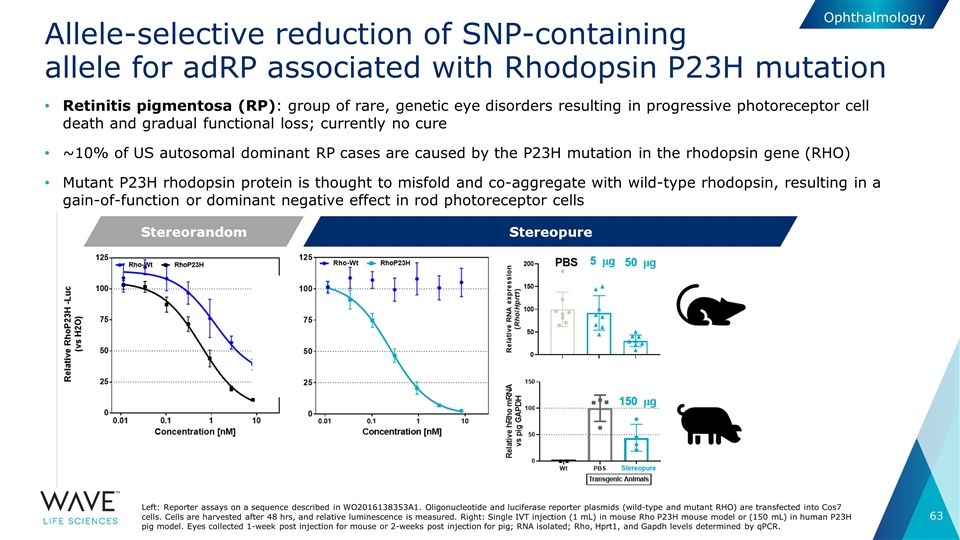
Allele-selective reduction of SNP-containing allele for adRP associated with Rhodopsin P23H mutation Left: Reporter assays on a sequence described in WO2016138353A1. Oligonucleotide and luciferase reporter plasmids (wild-type and mutant RHO) are transfected into Cos7 cells. Cells are harvested after 48 hrs, and relative luminescence is measured. Right: Single IVT injection (1 mL) in mouse Rho P23H mouse model or (150 mL) in human P23H pig model. Eyes collected 1-week post injection for mouse or 2-weeks post injection for pig; RNA isolated; Rho, Hprt1, and Gapdh levels determined by qPCR. Ophthalmology Retinitis pigmentosa (RP): group of rare, genetic eye disorders resulting in progressive photoreceptor cell death and gradual functional loss; currently no cure ~10% of US autosomal dominant RP cases are caused by the P23H mutation in the rhodopsin gene (RHO) Mutant P23H rhodopsin protein is thought to misfold and co-aggregate with wild-type rhodopsin, resulting in a gain-of-function or dominant negative effect in rod photoreceptor cells
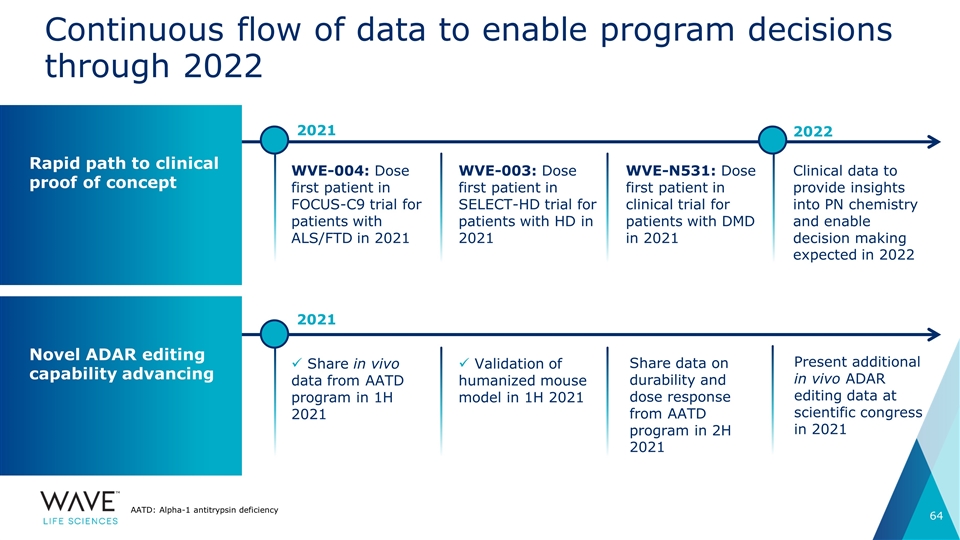
Validation of humanized mouse model in 1H 2021 WVE-004: Dose first patient in FOCUS-C9 trial for patients with ALS/FTD in 2021 Continuous flow of data to enable program decisions through 2022 WVE-003: Dose first patient in SELECT-HD trial for patients with HD in 2021 WVE-N531: Dose first patient in clinical trial for patients with DMD in 2021 Clinical data to provide insights into PN chemistry and enable decision making expected in 2022 Rapid path to clinical proof of concept Novel ADAR editing capability advancing Share in vivo data from AATD program in 1H 2021 Present additional in vivo ADAR editing data at scientific congress in 2021 AATD: Alpha-1 antitrypsin deficiency 2021 2022 2021 Share data on durability and dose response from AATD program in 2H 2021

Realizing a brighter future for people affected by genetic diseases For more information: Kate Rausch, Investor Relations krausch@wavelifesci.com 617.949.4827
































































