
Corporate Presentation August 8, 2024 Wave Life Sciences Exhibit 99.2

Forward-looking statements This document contains forward-looking statements. All statements other than statements of historical facts contained in this document, including statements regarding possible or assumed future results of operations, preclinical and clinical studies, business strategies, research and development plans, collaborations and partnerships, regulatory activities and timing thereof, competitive position, potential growth opportunities, use of proceeds and the effects of competition are forward-looking statements. These statements involve known and unknown risks, uncertainties and other important factors that may cause the actual results, performance or achievements of Wave Life Sciences Ltd. (the “Company”) to be materially different from any future results, performance or achievements expressed or implied by the forward-looking statements. In some cases, you can identify forward-looking statements by terms such as “may,” “will,” “should,” “expect,” “plan,” “aim,” “anticipate,” “could,” “intend,” “target,” “project,” “contemplate,” “believe,” “estimate,” “predict,” “potential” or “continue” or the negative of these terms or other similar expressions. The forward-looking statements in this presentation are only predictions. The Company has based these forward-looking statements largely on its current expectations and projections about future events and financial trends that it believes may affect the Company’s business, financial condition and results of operations. These forward-looking statements speak only as of the date of this presentation and are subject to a number of risks, uncertainties and assumptions, including those listed under Risk Factors in the Company’s Form 10-K and other filings with the SEC, some of which cannot be predicted or quantified and some of which are beyond the Company’s control. The events and circumstances reflected in the Company’s forward-looking statements may not be achieved or occur, and actual results could differ materially from those projected in the forward-looking statements. Moreover, the Company operates in a dynamic industry and economy. New risk factors and uncertainties may emerge from time to time, and it is not possible for management to predict all risk factors and uncertainties that the Company may face. Except as required by applicable law, the Company does not plan to publicly update or revise any forward-looking statements contained herein, whether as a result of any new information, future events, changed circumstances or otherwise.

AATD: Alpha-1 antitrypsin deficiency DMD: Duchenne muscular dystrophy HD: Huntington’s disease Building a leading RNA medicines company Multi-modal: RNA editing, RNAi, splicing, allele-selective silencing Best-in-class, clinically-validated oligonucleotide chemistry (PN, stereochemistry) Novel RNA medicines platform (PRISM®) WVE-003 in HD WVE-N531 in DMD WVE-007 in Obesity WVE-006 in AATD Strategic collaborations (GSK and Takeda) In-house GMP manufacturing Strong and broad IP Differentiated RNA medicines pipeline
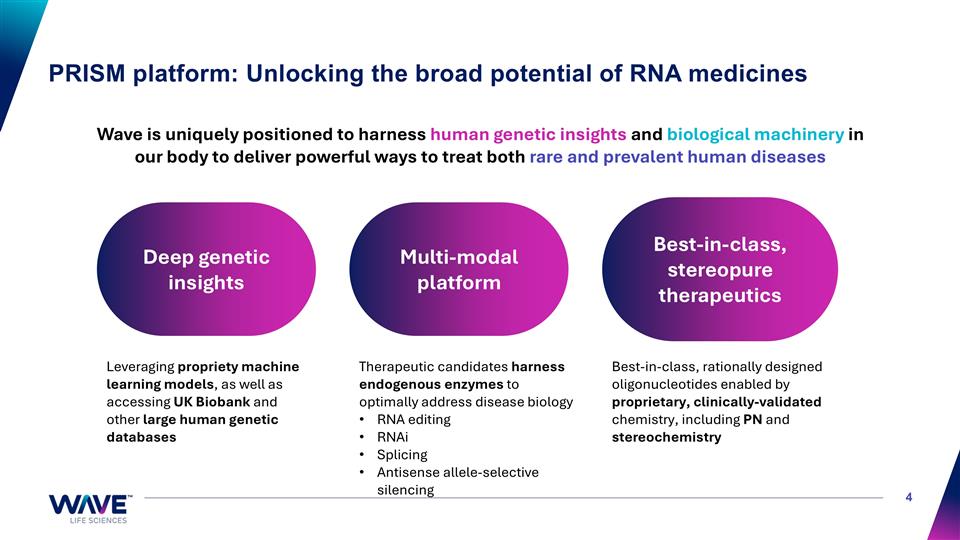
PRISM platform: Unlocking the broad potential of RNA medicines Wave is uniquely positioned to harness human genetic insights and biological machinery in our body to deliver powerful ways to treat both rare and prevalent human diseases Best-in-class, rationally designed oligonucleotides enabled by proprietary, clinically-validated chemistry, including PN and stereochemistry Therapeutic candidates harness endogenous enzymes to optimally address disease biology RNA editing RNAi Splicing Antisense allele-selective silencing Leveraging propriety machine learning models, as well as accessing UK Biobank and other large human genetic databases Best-in-class, stereopure therapeutics Deep genetic insights Multi-modal platform
Full list of Wave publications: https://ir.wavelifesciences.com/events-publications/publications Wave has driven foundational advances in nucleic acid chemistry to expand platform technologies and develop next generation of RNA therapeutics Further information can be found in recent platform publications Silencing (RNase H and Ago2) Splicing Editing

Full list of Wave publications: https://ir.wavelifesciences.com/events-publications/publications *mHTT reductions compared to placebo Proprietary chemistry continues to translate in clinic across modalities, enabling first-in-class and best-in-class therapies Preclinical publication Clinical translation Clinical study results 35% allele-selective mHTT silencing with single dose ✓ 46% allele-selective mHTT silencing with multi-dose, correlation with slowing of caudate atrophy ✓ ✓ Dystrophin data expected 3Q 2024 Novel base and sugar chemistry modifications Stereopure oligonucleotides Novel backbone modifications (including PN chemistry) Proprietary PRISM platform Therapeutic modalities Allele-selective silencing (WVE-003 for HD) Splicing (WVE-N531 for DMD) GalNAc-RNA editing (WVE-006 for AATD) GalNAc-RNAi (WVE-007 for obesity) ✓ ✓ ✓ 53% exon skipping, 42 µg/g muscle tissue concentrations in 6 weeks ✓ Proof-of-mechanism data expected 4Q 2024 Clinical trial initiation expected 1Q 2025 RestorAATion study completion
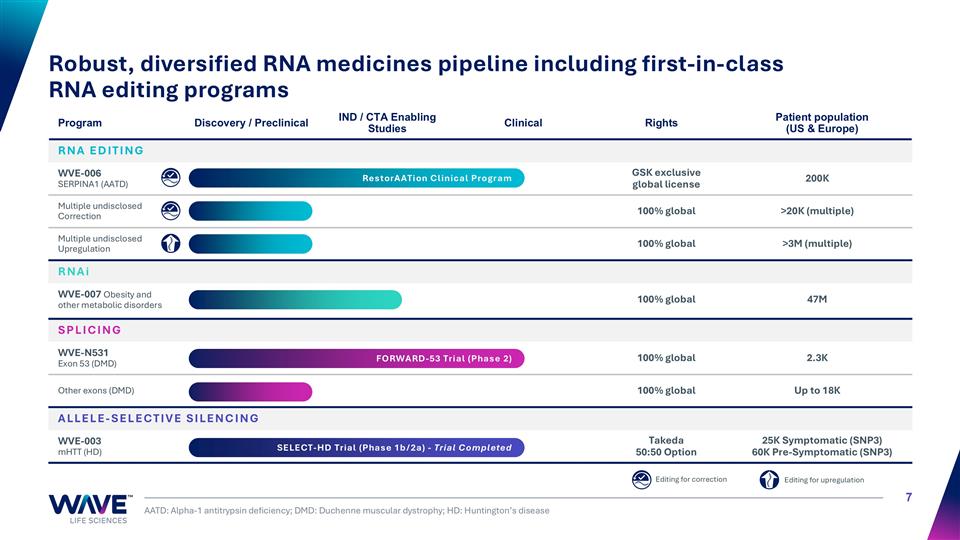
SPLICING WVE-N531 Exon 53 (DMD) 100% global 2.3K Other exons (DMD) 100% global Up to 18K ALLELE-SELECTIVE SILENCING WVE-003 mHTT (HD) Takeda 50:50 Option 25K Symptomatic (SNP3) 60K Pre-Symptomatic (SNP3) Robust, diversified RNA medicines pipeline including first-in-class RNA editing programs AATD: Alpha-1 antitrypsin deficiency; DMD: Duchenne muscular dystrophy; HD: Huntington’s disease Program Discovery / Preclinical IND / CTA Enabling Studies Clinical Rights Patient population (US & Europe) RNA EDITING WVE-006 SERPINA1 (AATD) GSK exclusive global license 200K Multiple undisclosed Correction 100% global >20K (multiple) Multiple undisclosed Upregulation 100% global >3M (multiple) RNAi WVE-007 Obesity and other metabolic disorders 100% global 47M Editing for correction Editing for upregulation FORWARD-53 Trial (Phase 2) SELECT-HD Trial (Phase 1b/2a) - Trial Completed RestorAATion Clinical Program

WVE-006 + AIMers RNA editing Alpha-1 antitrypsin deficiency (AATD)
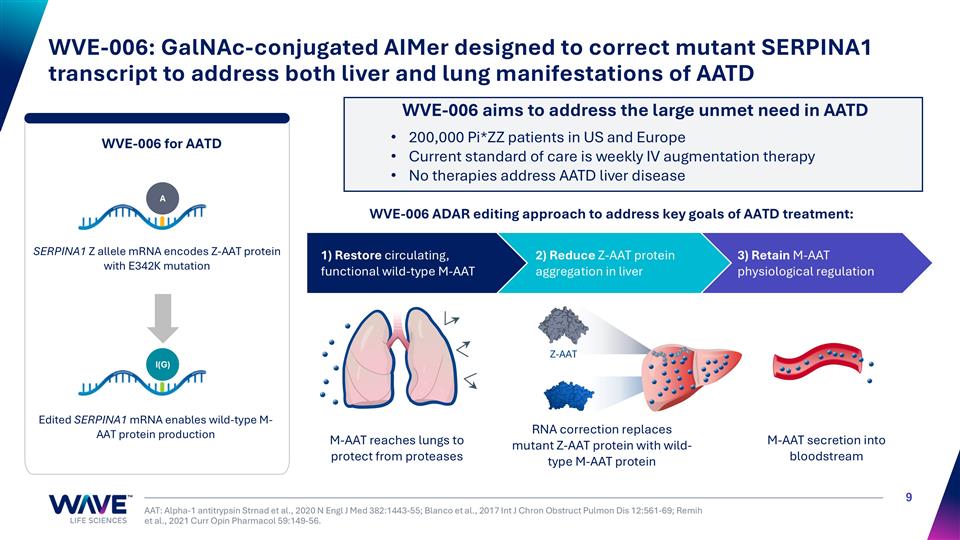
WVE-006: GalNAc-conjugated AIMer designed to correct mutant SERPINA1 transcript to address both liver and lung manifestations of AATD AAT: Alpha-1 antitrypsin Strnad et al., 2020 N Engl J Med 382:1443-55; Blanco et al., 2017 Int J Chron Obstruct Pulmon Dis 12:561-69; Remih et al., 2021 Curr Opin Pharmacol 59:149-56. 3) Retain M-AAT physiological regulation 2) Reduce Z-AAT protein aggregation in liver WVE-006 ADAR editing approach to address key goals of AATD treatment: 1) Restore circulating, functional wild-type M-AAT I(G) A SERPINA1 Z allele mRNA encodes Z-AAT protein with E342K mutation Edited SERPINA1 mRNA enables wild-type M-AAT protein production WVE-006 for AATD WVE-006 aims to address the large unmet need in AATD M-AAT reaches lungs to protect from proteases M-AAT secretion into bloodstream RNA correction replaces mutant Z-AAT protein with wild-type M-AAT protein Z-AAT 200,000 Pi*ZZ patients in US and Europe Current standard of care is weekly IV augmentation therapy No therapies address AATD liver disease
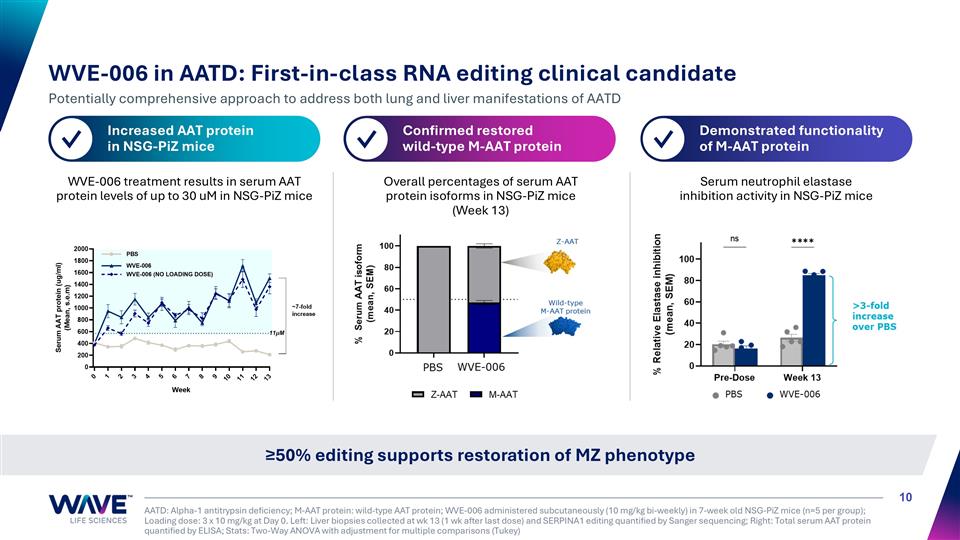
WVE-006 treatment results in serum AAT protein levels of up to 30 uM in NSG-PiZ mice Overall percentages of serum AAT protein isoforms in NSG-PiZ mice (Week 13) Serum neutrophil elastase inhibition activity in NSG-PiZ mice AATD: Alpha-1 antitrypsin deficiency; M-AAT protein: wild-type AAT protein; WVE-006 administered subcutaneously (10 mg/kg bi-weekly) in 7-week old NSG-PiZ mice (n=5 per group); Loading dose: 3 x 10 mg/kg at Day 0. Left: Liver biopsies collected at wk 13 (1 wk after last dose) and SERPINA1 editing quantified by Sanger sequencing; Right: Total serum AAT protein quantified by ELISA; Stats: Two-Way ANOVA with adjustment for multiple comparisons (Tukey) ≥50% editing supports restoration of MZ phenotype WVE-006 in AATD: First-in-class RNA editing clinical candidate Potentially comprehensive approach to address both lung and liver manifestations of AATD Increased AAT protein in NSG-PiZ mice ✓ Confirmed restored wild-type M-AAT protein ✓ Demonstrated functionality of M-AAT protein ✓
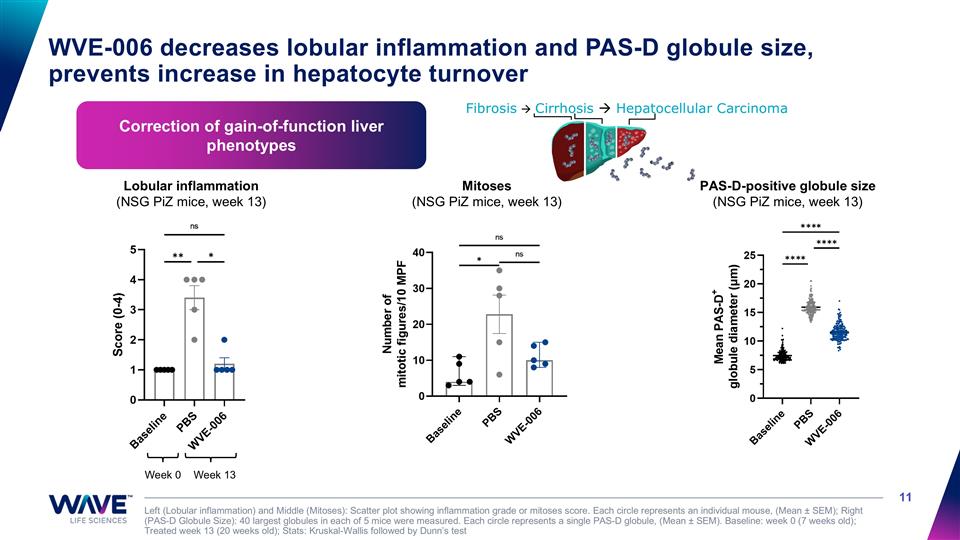
WVE-006 decreases lobular inflammation and PAS-D globule size, prevents increase in hepatocyte turnover Left (Lobular inflammation) and Middle (Mitoses): Scatter plot showing inflammation grade or mitoses score. Each circle represents an individual mouse, (Mean ± SEM); Right (PAS-D Globule Size): 40 largest globules in each of 5 mice were measured. Each circle represents a single PAS-D globule, (Mean ± SEM). Baseline: week 0 (7 weeks old); Treated week 13 (20 weeks old); Stats: Kruskal-Wallis followed by Dunn’s test Mitoses (NSG PiZ mice, week 13) Fibrosis à Cirrhosis à Hepatocellular Carcinoma Correction of gain-of-function liver phenotypes Lobular inflammation (NSG PiZ mice, week 13) Week 0 Week 13 PAS-D-positive globule size (NSG PiZ mice, week 13)
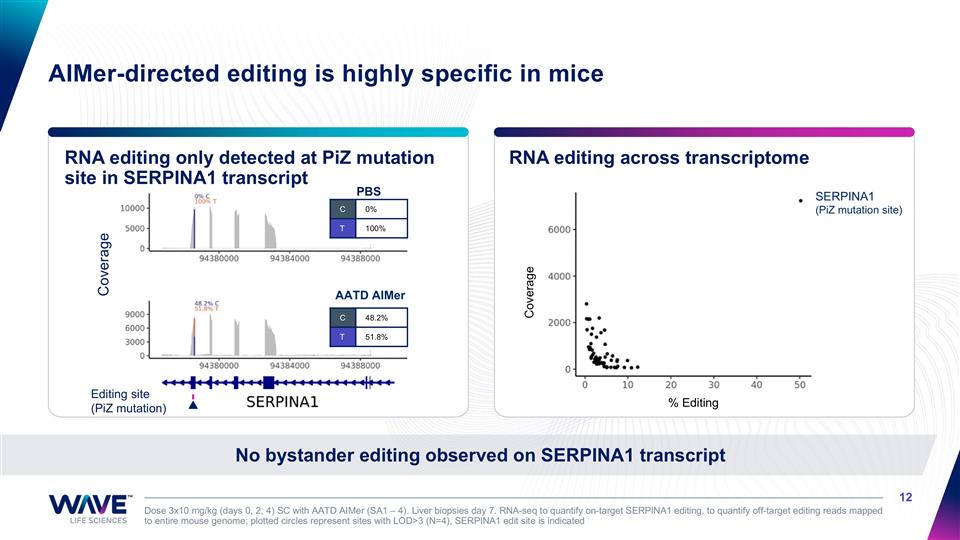
RNA editing only detected at PiZ mutation site in SERPINA1 transcript RNA editing across transcriptome Dose 3x10 mg/kg (days 0, 2, 4) SC with AATD AIMer (SA1 – 4). Liver biopsies day 7. RNA-seq to quantify on-target SERPINA1 editing, to quantify off-target editing reads mapped to entire mouse genome; plotted circles represent sites with LOD>3 (N=4), SERPINA1 edit site is indicated No bystander editing observed on SERPINA1 transcript AIMer-directed editing is highly specific in mice SERPINA1 (PiZ mutation site) Coverage PBS AATD AIMer C 48.2% T 51.8% C 0% T 100% Editing site (PiZ mutation) Coverage % Editing
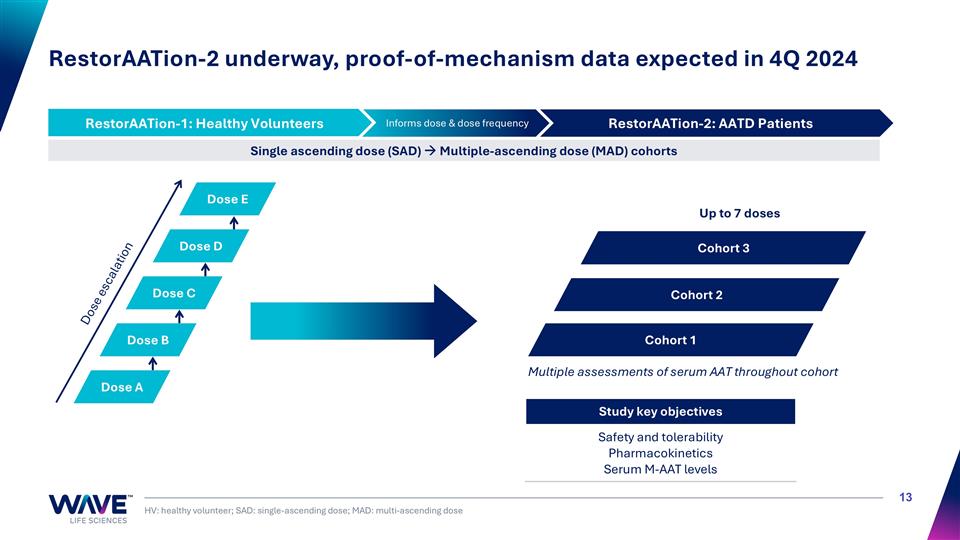
RestorAATion-2 underway, proof-of-mechanism data expected in 4Q 2024 HV: healthy volunteer; SAD: single-ascending dose; MAD: multi-ascending dose RestorAATion-2: AATD Patients Informs dose & dose frequency RestorAATion-1: Healthy Volunteers Single ascending dose (SAD) à Multiple-ascending dose (MAD) cohorts Dose escalation Study key objectives Safety and tolerability Pharmacokinetics Serum M-AAT levels Multiple assessments of serum AAT throughout cohort Dose A Cohort 1 Up to 7 doses Dose B Dose C Dose D Dose E Cohort 2 Cohort 3 RestorAATion-1: Healthy Volunteers
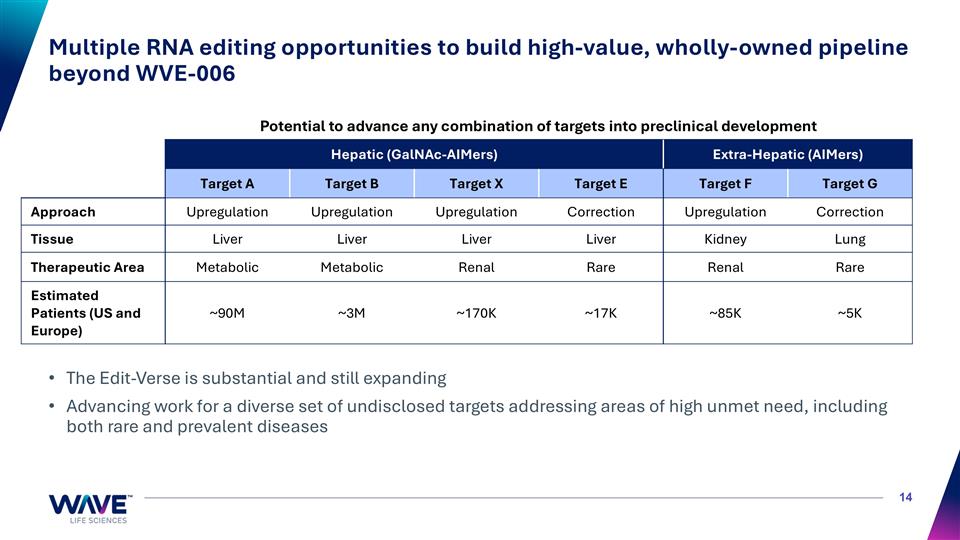
Multiple RNA editing opportunities to build high-value, wholly-owned pipeline beyond WVE-006 The Edit-Verse is substantial and still expanding Advancing work for a diverse set of undisclosed targets addressing areas of high unmet need, including both rare and prevalent diseases Hepatic (GalNAc-AIMers) Extra-Hepatic (AIMers) Target A Target B Target X Target E Target F Target G Approach Upregulation Upregulation Upregulation Correction Upregulation Correction Tissue Liver Liver Liver Liver Kidney Lung Therapeutic Area Metabolic Metabolic Renal Rare Renal Rare Estimated Patients (US and Europe) ~90M ~3M ~170K ~17K ~85K ~5K Potential to advance any combination of targets into preclinical development
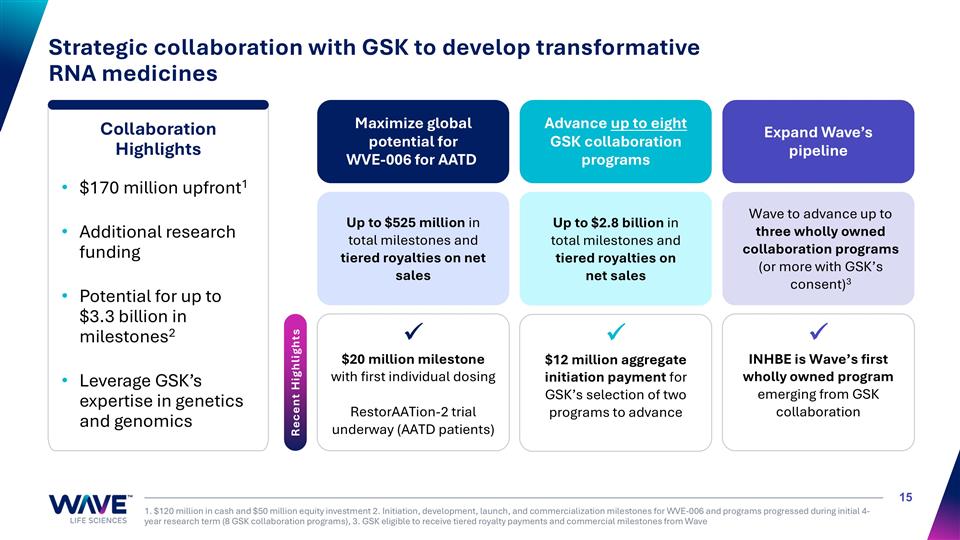
Strategic collaboration with GSK to develop transformative RNA medicines 1. $120 million in cash and $50 million equity investment 2. Initiation, development, launch, and commercialization milestones for WVE-006 and programs progressed during initial 4-year research term (8 GSK collaboration programs), 3. GSK eligible to receive tiered royalty payments and commercial milestones from Wave Maximize global potential for WVE-006 for AATD Advance up to eight GSK collaboration programs Up to $525 million in total milestones and tiered royalties on net sales Up to $2.8 billion in total milestones and tiered royalties on net sales Wave to advance up to three wholly owned collaboration programs (or more with GSK’s consent)3 Expand Wave’s pipeline Recent Highlights ü $20 million milestone with first individual dosing RestorAATion-2 trial underway (AATD patients) Collaboration Highlights $170 million upfront1 Additional research funding Potential for up to $3.3 billion in milestones2 Leverage GSK’s expertise in genetics and genomics ü $12 million aggregate initiation payment for GSK’s selection of two programs to advance ü INHBE is Wave’s first wholly owned program emerging from GSK collaboration

WVE-007 (INHBE program) GalNAc-siRNA silencing Obesity and other metabolic disorders
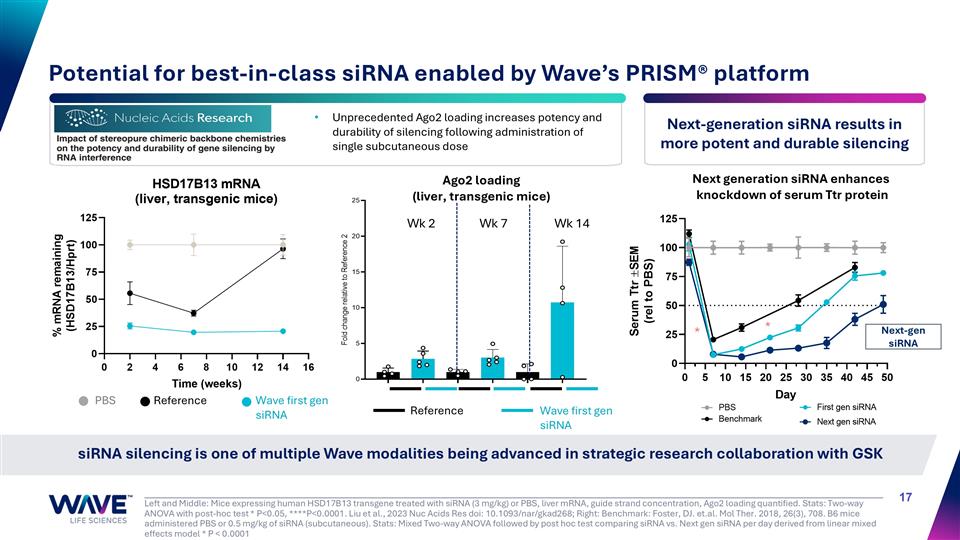
Left and Middle: Mice expressing human HSD17B13 transgene treated with siRNA (3 mg/kg) or PBS, liver mRNA, guide strand concentration, Ago2 loading quantified. Stats: Two-way ANOVA with post-hoc test * P<0.05, ****P<0.0001. Liu et al., 2023 Nuc Acids Res doi: 10.1093/nar/gkad268; Right: Benchmark: Foster, DJ. et.al. Mol Ther. 2018, 26(3), 708. B6 mice administered PBS or 0.5 mg/kg of siRNA (subcutaneous). Stats: Mixed Two-way ANOVA followed by post hoc test comparing siRNA vs. Next gen siRNA per day derived from linear mixed effects model * P < 0.0001 siRNA silencing is one of multiple Wave modalities being advanced in strategic research collaboration with GSK Potential for best-in-class siRNA enabled by Wave’s PRISM® platform Wk 2 Wk 14 Wk 7 Reference Wave first gen siRNA * * Ago2 loading (liver, transgenic mice) Wave first gen siRNA Reference PBS Unprecedented Ago2 loading increases potency and durability of silencing following administration of single subcutaneous dose Next-gen siRNA Next generation siRNA enhances knockdown of serum Ttr protein Next-generation siRNA results in more potent and durable silencing
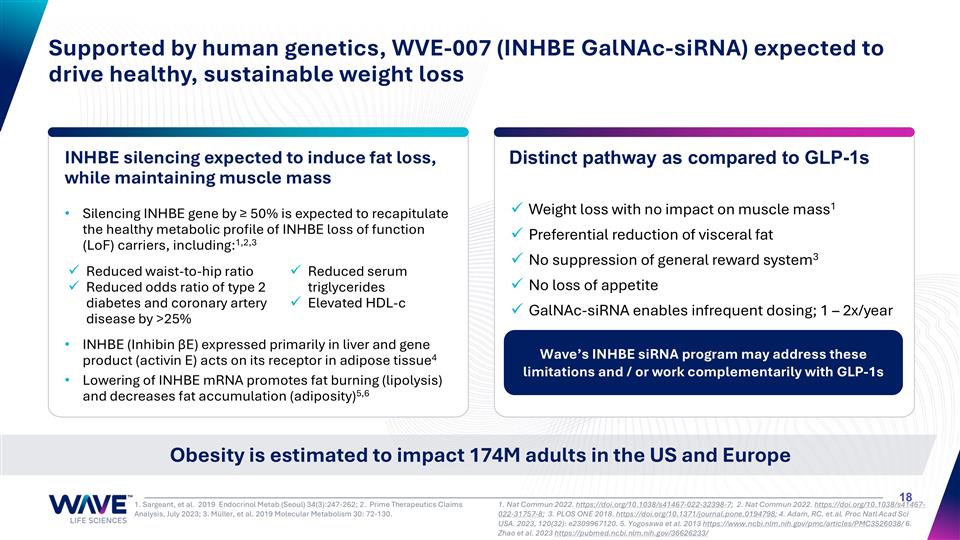
Silencing INHBE gene by ≥ 50% is expected to recapitulate the healthy metabolic profile of INHBE loss of function (LoF) carriers, including:1,2,3 INHBE (Inhibin βE) expressed primarily in liver and gene product (activin E) acts on its receptor in adipose tissue4 Lowering of INHBE mRNA promotes fat burning (lipolysis) and decreases fat accumulation (adiposity)5,6 INHBE silencing expected to induce fat loss, while maintaining muscle mass Obesity is estimated to impact 174M adults in the US and Europe Supported by human genetics, WVE-007 (INHBE GalNAc-siRNA) expected to drive healthy, sustainable weight loss 1. Nat Commun 2022. https://doi.org/10.1038/s41467-022-32398-7; 2. Nat Commun 2022. https://doi.org/10.1038/s41467-022-31757-8; 3. PLOS ONE 2018. https://doi.org/10.1371/journal.pone.0194798; 4. Adam, RC. et.al. Proc Natl Acad Sci USA. 2023, 120(32): e2309967120. 5. Yogosawa et al. 2013 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3526038/ 6. Zhao et al. 2023 https://pubmed.ncbi.nlm.nih.gov/36626233/ 1. Sargeant, et al. 2019 Endocrinol Metab (Seoul) 34(3):247-262; 2. Prime Therapeutics Claims Analysis, July 2023; 3. Müller, et al. 2019 Molecular Metabolism 30: 72-130. Reduced waist-to-hip ratio Reduced odds ratio of type 2 diabetes and coronary artery disease by >25% Reduced serum triglycerides Elevated HDL-c Distinct pathway as compared to GLP-1s Weight loss with no impact on muscle mass1 Preferential reduction of visceral fat No suppression of general reward system3 No loss of appetite GalNAc-siRNA enables infrequent dosing; 1 – 2x/year Wave’s INHBE siRNA program may address these limitations and / or work complementarily with GLP-1s

Highly potent (ED50 < 1mg/kg) and durable silencing following one, low-single-digit dose, supporting every-six-month or annual dosing Monotherapy: Weight loss similar to semaglutide with no loss of muscle mass and a reduction in fat mass, with preferential effect to the visceral fat (consistent with profile of INHBE LoF carriers in human genetics) Combination with GLP-1s: When administered in combination with semaglutide, a single dose of Wave’s INHBE GalNAc-siRNA doubled the weight loss observed with semaglutide alone and this effect was sustained throughout the duration of the study Maintenance: Curtailed rebound weight gain upon cessation of semaglutide INHBE program: Data from DIO mouse model supports best-in-class profile and potential use of WVE-007 in multiple treatment settings WVE-007 has Wave’s next generation siRNA format and best-in-class profile with infrequent dosing Expect to initiate clinical trial for WVE-007 in 1Q 2025

WVE-N531 Splicing Duchenne muscular dystrophy
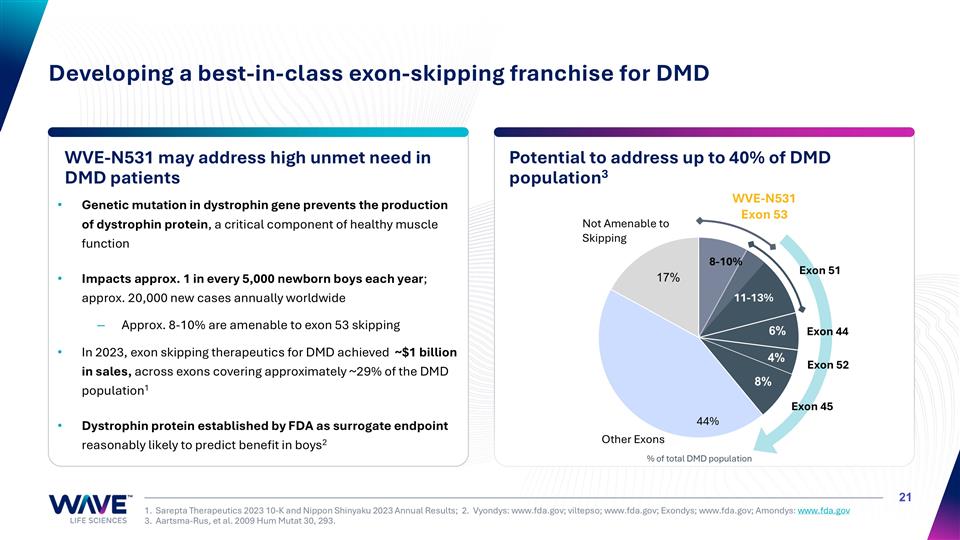
Genetic mutation in dystrophin gene prevents the production of dystrophin protein, a critical component of healthy muscle function Impacts approx. 1 in every 5,000 newborn boys each year; approx. 20,000 new cases annually worldwide Approx. 8-10% are amenable to exon 53 skipping In 2023, exon skipping therapeutics for DMD achieved ~$1 billion in sales, across exons covering approximately ~29% of the DMD population1 Dystrophin protein established by FDA as surrogate endpoint reasonably likely to predict benefit in boys2 WVE-N531 may address high unmet need in DMD patients Potential to address up to 40% of DMD population3 1. Sarepta Therapeutics 2023 10-K and Nippon Shinyaku 2023 Annual Results; 2. Vyondys: www.fda.gov; viltepso; www.fda.gov; Exondys; www.fda.gov; Amondys: www.fda.gov 3. Aartsma-Rus, et al. 2009 Hum Mutat 30, 293. Developing a best-in-class exon-skipping franchise for DMD Exon 45 Exon 44 Exon 52 WVE-N531 Exon 53 Exon 51 Not Amenable to Skipping 11-13% 8-10% 44% % of total DMD population
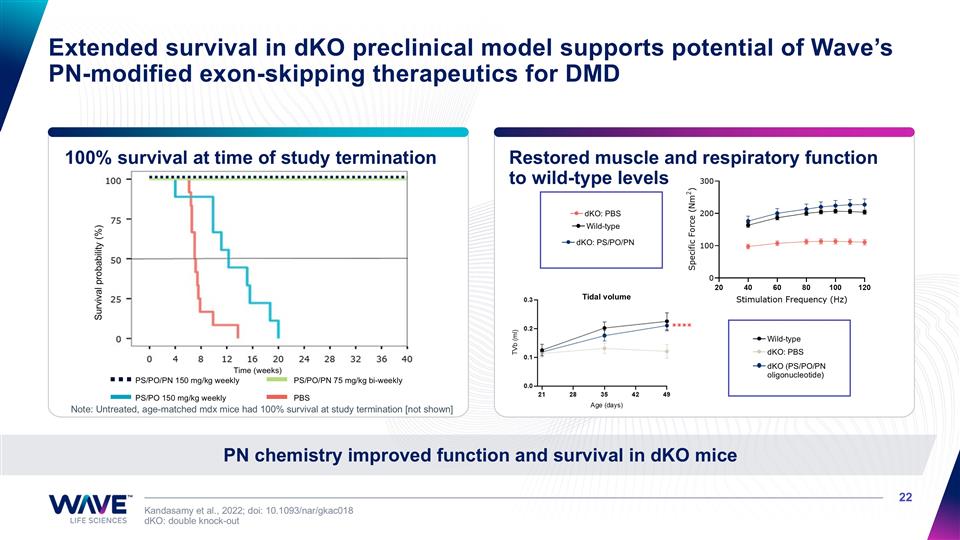
100% survival at time of study termination Restored muscle and respiratory function to wild-type levels Kandasamy et al., 2022; doi: 10.1093/nar/gkac018 dKO: double knock-out PN chemistry improved function and survival in dKO mice Extended survival in dKO preclinical model supports potential of Wave’s PN-modified exon-skipping therapeutics for DMD Wild-type dKO: PBS dKO: PS/PO/PN Wild-type dKO: PBS dKO (PS/PO/PN oligonucleotide) PS/PO/PN 150 mg/kg weekly PS/PO/PN 75 mg/kg bi-weekly PS/PO 150 mg/kg weekly PBS Note: Untreated, age-matched mdx mice had 100% survival at study termination [not shown] Tidal volume Age (days) TVb (ml) Survival probability (%) Time (weeks)
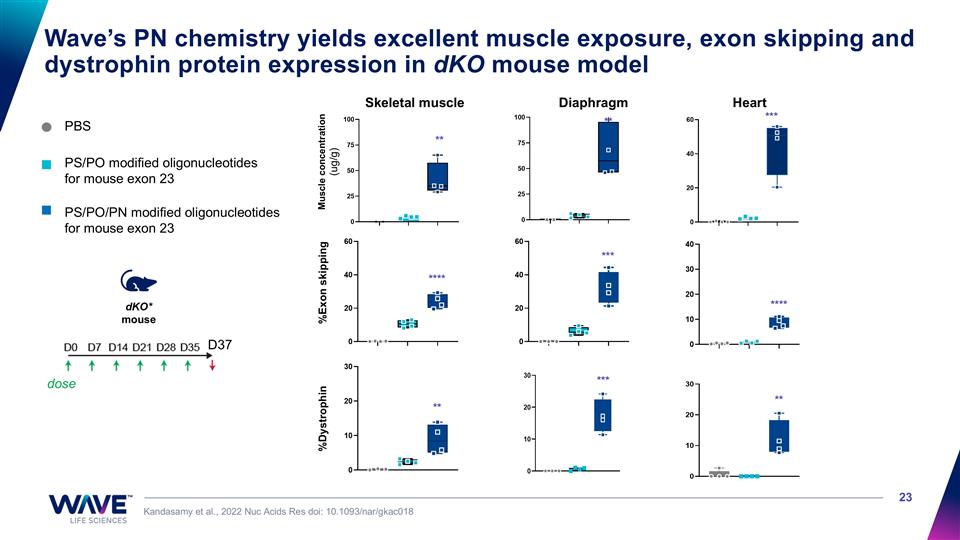
PBS PS/PO/PN modified oligonucleotides for mouse exon 23 Wave’s PN chemistry yields excellent muscle exposure, exon skipping and dystrophin protein expression in dKO mouse model Kandasamy et al., 2022 Nuc Acids Res doi: 10.1093/nar/gkac018 dose dKO* mouse D37 %Dystrophin Skeletal muscle Diaphragm Heart %Exon skipping Muscle concentration (ug/g) **** *** **** ** *** ** ** ** *** PS/PO modified oligonucleotides for mouse exon 23
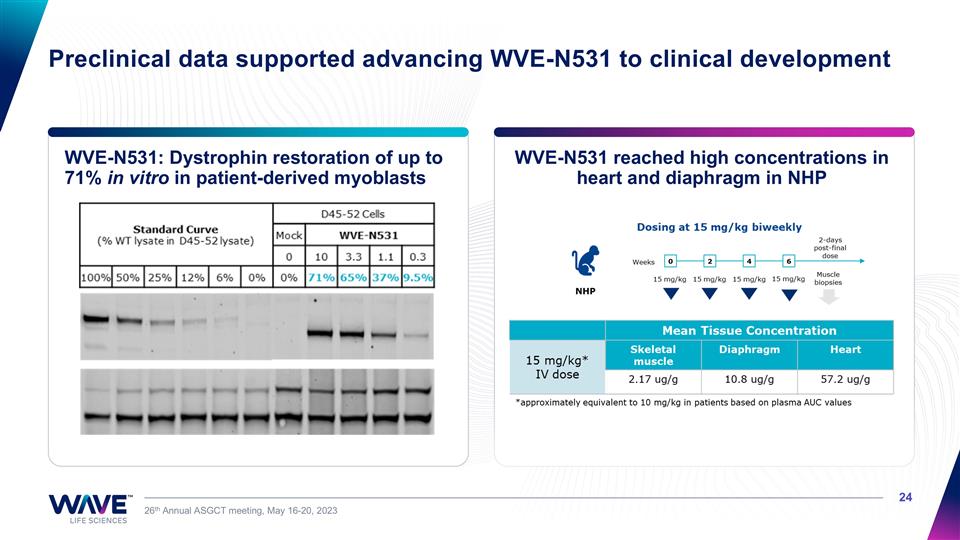
Preclinical data supported advancing WVE-N531 to clinical development WVE-N531 reached high concentrations in heart and diaphragm in NHP 26th Annual ASGCT meeting, May 16-20, 2023 WVE-N531: Dystrophin restoration of up to 71% in vitro in patient-derived myoblasts
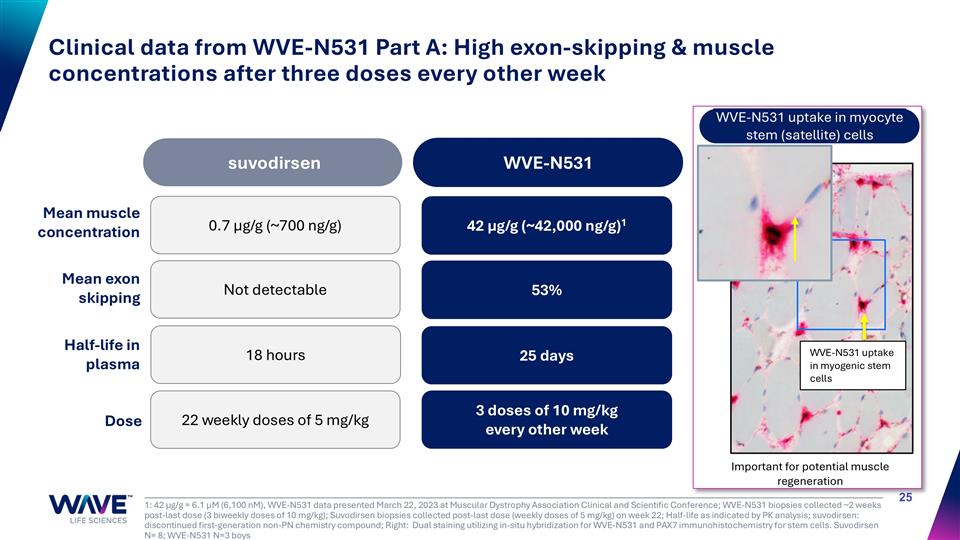
1: 42 µg/g = 6.1 µM (6,100 nM), WVE-N531 data presented March 22, 2023 at Muscular Dystrophy Association Clinical and Scientific Conference; WVE-N531 biopsies collected ~2 weeks post-last dose (3 biweekly doses of 10 mg/kg); Suvodirsen biopsies collected post-last dose (weekly doses of 5 mg/kg) on week 22; Half-life as indicated by PK analysis; suvodirsen: discontinued first-generation non-PN chemistry compound; Right: Dual staining utilizing in-situ hybridization for WVE-N531 and PAX7 immunohistochemistry for stem cells. Suvodirsen N= 8; WVE-N531 N=3 boys Clinical data from WVE-N531 Part A: High exon-skipping & muscle concentrations after three doses every other week WVE-N531 uptake in myogenic stem cells Important for potential muscle regeneration Mean muscle concentration Mean exon skipping Half-life in plasma Dose suvodirsen WVE-N531 0.7 µg/g (~700 ng/g) Not detectable 18 hours 22 weekly doses of 5 mg/kg 42 µg/g (~42,000 ng/g)1 53% 25 days 3 doses of 10 mg/kg every other week WVE-N531 uptake in myocyte stem (satellite) cells

Design of FORWARD-53: Phase 2, open-label, 10 mg/kg every other week Endpoints: Dystrophin (powered for >5% of normal), safety/tolerability, pharmacokinetics, digital and functional assessments (incl. NSAA and others) Muscle biopsies to assess dystrophin expression Fully enrolled (n=11) and dosing underway IV: intravenous; NSAA: North star ambulatory assessment Potentially registrational 24-week dystrophin expression data are expected in 3Q 2024 Advancing FORWARD-53, a potentially registrational Phase 2 clinical trial of WVE-N531 in DMD (Exon 53) Screening Every other week IV dosing Functional assessment Biopsy after 24 weeks of treatment Functional assessment Biopsy after 48 weeks of treatment Functional assessment Safety Follow-up

WVE-003 Allele-selective silencing Huntington’s Disease
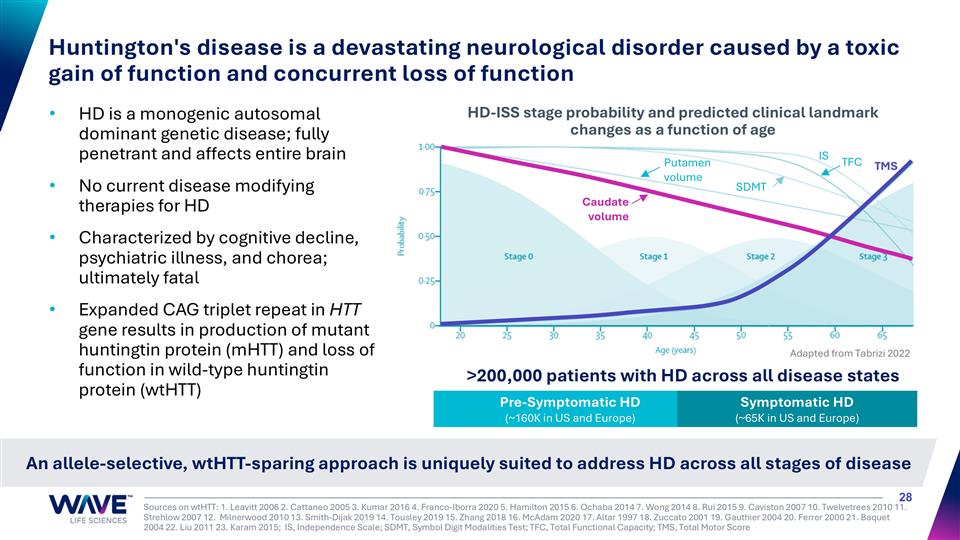
HD is a monogenic autosomal dominant genetic disease; fully penetrant and affects entire brain No current disease modifying therapies for HD Characterized by cognitive decline, psychiatric illness, and chorea; ultimately fatal Expanded CAG triplet repeat in HTT gene results in production of mutant huntingtin protein (mHTT) and loss of function in wild-type huntingtin protein (wtHTT) Huntington's disease is a devastating neurological disorder caused by a toxic gain of function and concurrent loss of function Pre-Symptomatic HD (~160K in US and Europe) Symptomatic HD (~65K in US and Europe) Putamen volume Caudate volume IS TFC TMS SDMT Adapted from Tabrizi 2022 HD-ISS stage probability and predicted clinical landmark changes as a function of age Sources on wtHTT: 1. Leavitt 2006 2. Cattaneo 2005 3. Kumar 2016 4. Franco-Iborra 2020 5. Hamilton 2015 6. Ochaba 2014 7. Wong 2014 8. Rui 2015 9. Caviston 2007 10. Twelvetrees 2010 11. Strehlow 2007 12. Milnerwood 2010 13. Smith-Dijak 2019 14. Tousley 2019 15. Zhang 2018 16. McAdam 2020 17. Altar 1997 18. Zuccato 2001 19. Gauthier 2004 20. Ferrer 2000 21. Baquet 2004 22. Liu 2011 23. Karam 2015; IS, Independence Scale; SDMT, Symbol Digit Modalities Test; TFC, Total Functional Capacity; TMS, Total Motor Score An allele-selective, wtHTT-sparing approach is uniquely suited to address HD across all stages of disease >200,000 patients with HD across all disease states
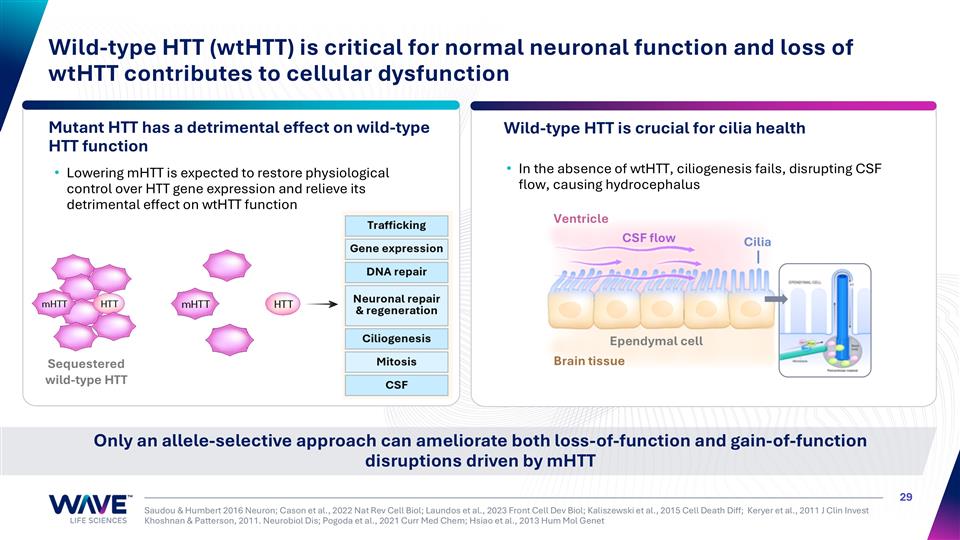
Saudou & Humbert 2016 Neuron; Cason et al., 2022 Nat Rev Cell Biol; Laundos et al., 2023 Front Cell Dev Biol; Kaliszewski et al., 2015 Cell Death Diff; Keryer et al., 2011 J Clin Invest Khoshnan & Patterson, 2011. Neurobiol Dis; Pogoda et al., 2021 Curr Med Chem; Hsiao et al., 2013 Hum Mol Genet Wild-type HTT (wtHTT) is critical for normal neuronal function and loss of wtHTT contributes to cellular dysfunction Lowering mHTT is expected to restore physiological control over HTT gene expression and relieve its detrimental effect on wtHTT function In the absence of wtHTT, ciliogenesis fails, disrupting CSF flow, causing hydrocephalus Mutant HTT has a detrimental effect on wild-type HTT function Wild-type HTT is crucial for cilia health Only an allele-selective approach can ameliorate both loss-of-function and gain-of-function disruptions driven by mHTT Brain tissue Ventricle Cilia CSF flow Ependymal cell Sequestered wild-type HTT
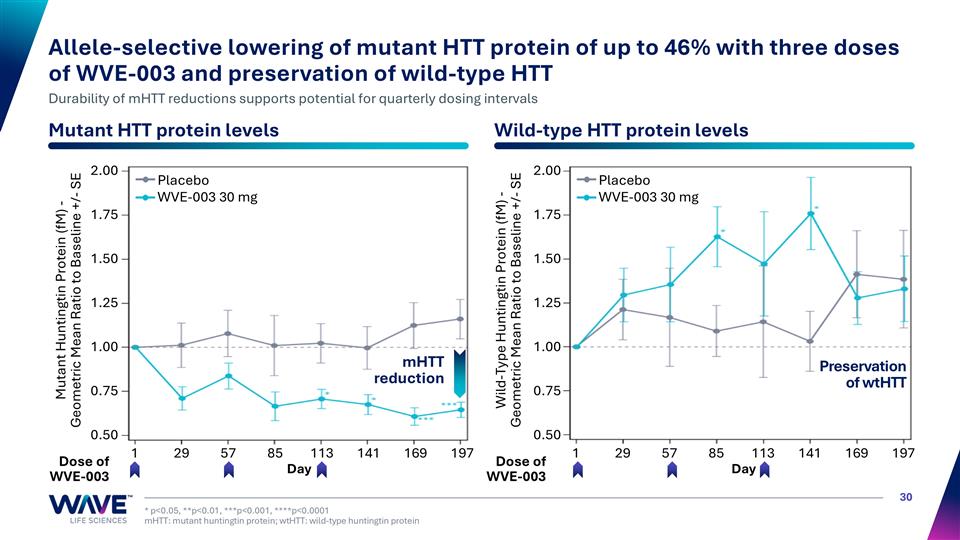
* p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 mHTT: mutant huntingtin protein; wtHTT: wild-type huntingtin protein Allele-selective lowering of mutant HTT protein of up to 46% with three doses of WVE-003 and preservation of wild-type HTT Durability of mHTT reductions supports potential for quarterly dosing intervals Mutant HTT protein levels Wild-type HTT protein levels Placebo WVE-003 30 mg Mutant Huntingtin Protein (fM) - Geometric Mean Ratio to Baseline +/- SE Wild-Type Huntingtin Protein (fM) - Geometric Mean Ratio to Baseline +/- SE Dose of WVE-003 Dose of WVE-003 Preservation of wtHTT mHTT reduction Day Day 2.00 1.75 1.50 1.25 1.00 0.75 0.50 2.00 1.75 1.50 1.25 1.00 0.75 0.50 1 29 57 85 113 141 169 197 1 29 57 85 113 141 169 197 Placebo WVE-003 30 mg
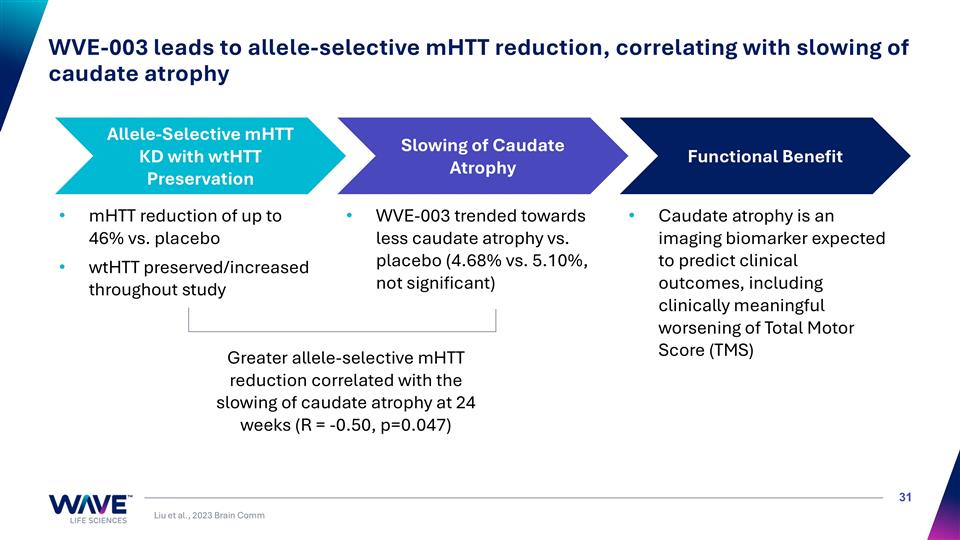
WVE-003 leads to allele-selective mHTT reduction, correlating with slowing of caudate atrophy Allele-Selective mHTT KD with wtHTT Preservation Slowing of Caudate Atrophy Functional Benefit mHTT reduction of up to 46% vs. placebo wtHTT preserved/increased throughout study Caudate atrophy is an imaging biomarker expected to predict clinical outcomes, including clinically meaningful worsening of Total Motor Score (TMS) WVE-003 trended towards less caudate atrophy vs. placebo (4.68% vs. 5.10%, not significant) Greater allele-selective mHTT reduction correlated with the slowing of caudate atrophy at 24 weeks (R = -0.50, p=0.047) Liu et al., 2023 Brain Comm
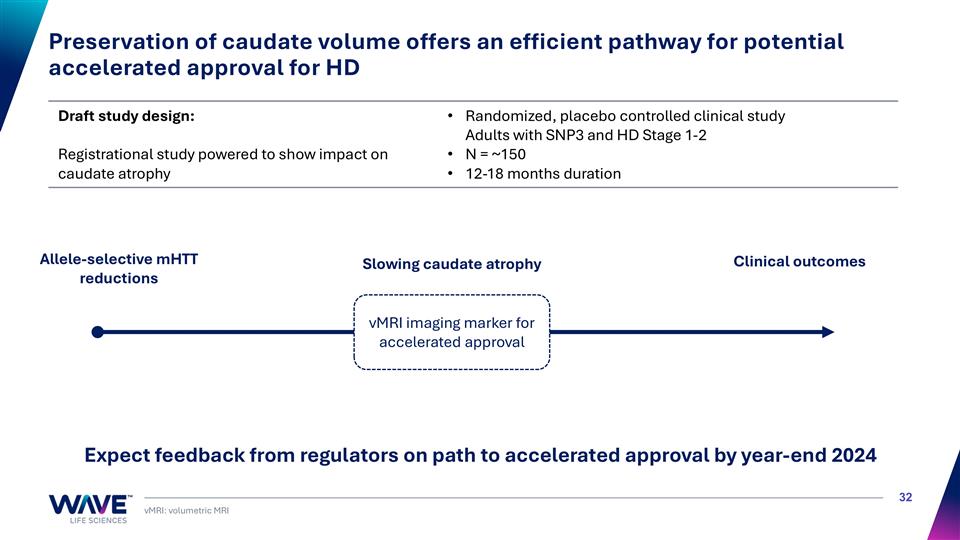
Draft study design: Registrational study powered to show impact on caudate atrophy Randomized, placebo controlled clinical study Adults with SNP3 and HD Stage 1-2 N = ~150 12-18 months duration vMRI: volumetric MRI Expect feedback from regulators on path to accelerated approval by year-end 2024 Allele-selective mHTT reductions Slowing caudate atrophy Preservation of caudate volume offers an efficient pathway for potential accelerated approval for HD Clinical outcomes vMRI imaging marker for accelerated approval

Anticipated upcoming milestones
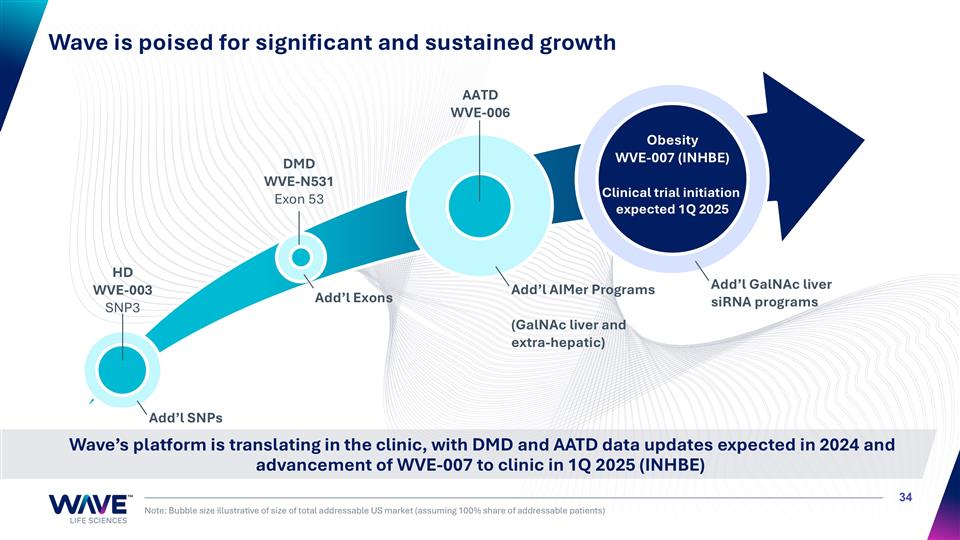
Wave is poised for significant and sustained growth Note: Bubble size illustrative of size of total addressable US market (assuming 100% share of addressable patients) AATD WVE-006 DMD WVE-N531 Exon 53 HD WVE-003 SNP3 Add’l AIMer Programs (GalNAc liver and extra-hepatic) Wave’s platform is translating in the clinic, with DMD and AATD data updates expected in 2024 and advancement of WVE-007 to clinic in 1Q 2025 (INHBE) Obesity WVE-007 (INHBE) Clinical trial initiation expected 1Q 2025 Add’l GalNAc liver siRNA programs
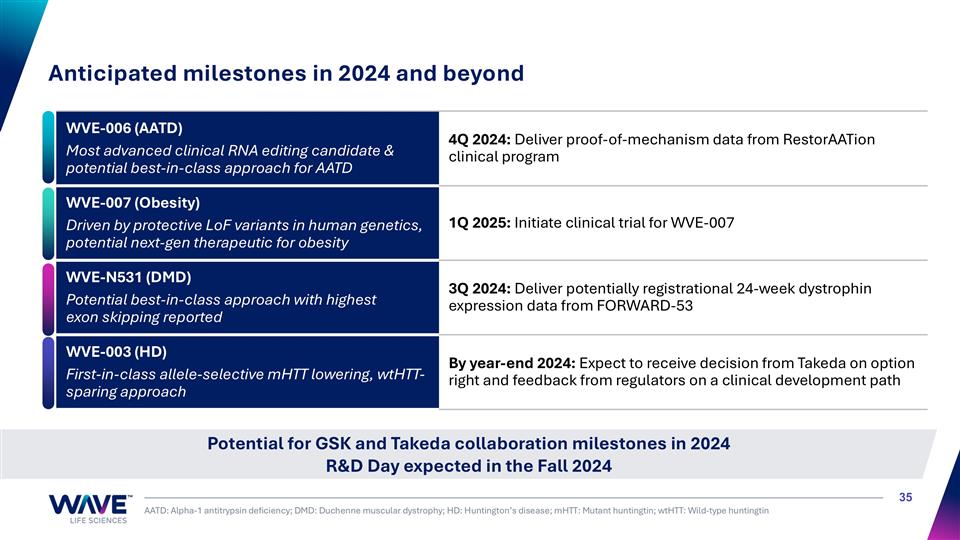
Potential for GSK and Takeda collaboration milestones in 2024 R&D Day expected in the Fall 2024 Anticipated milestones in 2024 and beyond AATD: Alpha-1 antitrypsin deficiency; DMD: Duchenne muscular dystrophy; HD: Huntington’s disease; mHTT: Mutant huntingtin; wtHTT: Wild-type huntingtin WVE-006 (AATD) Most advanced clinical RNA editing candidate & potential best-in-class approach for AATD 4Q 2024: Deliver proof-of-mechanism data from RestorAATion clinical program WVE-007 (Obesity) Driven by protective LoF variants in human genetics, potential next-gen therapeutic for obesity 1Q 2025: Initiate clinical trial for WVE-007 WVE-N531 (DMD) Potential best-in-class approach with highest exon skipping reported 3Q 2024: Deliver potentially registrational 24-week dystrophin expression data from FORWARD-53 WVE-003 (HD) First-in-class allele-selective mHTT lowering, wtHTT-sparing approach By year-end 2024: Expect to receive decision from Takeda on option right and feedback from regulators on a clinical development path

For questions contact: investorrelations@wavelifesci.com



































