
43rd Annual J.P. Morgan Healthcare Conference Nasdaq: MGX January 2025 Unlocking 4 Billion Years of Microbial Evolution to Create Curative Genetic Medicines

Forward Looking Statements This presentation includes forward-looking statements, including forward-looking statements within the meaning of the Private Securities Litigation Reform Act of 1995. All statements other than statements of historical facts contained in this presentation are forward looking statements, including statements regarding our cash runway, strategy and plans, industry environment, potential growth opportunities, and the therapeutic potential of our programs. The words “believe,” “may,” “will,” “estimate,” “continue,” “anticipate,” “ design,” “expect,” “could,” “plan,” “potential,” “predict,” “seek,” “should,” “would,” or the negative version of these words and similar expressions are intended to identify forward-looking statements. We have based these forward-looking statements on our current expectations and projections about future events and trends that we believe may affect our financial condition, results of operations, strategy, short and long term business operations and objectives, and financial needs. These forward-looking statements are subject to a number of risks, uncertainties and assumptions, including but not limited to, our ability to develop and advance our programs and product candidates, our ability to maintain and establish collaborations or strategic partnerships, our regulatory approvals and filings, and other risks, uncertainties and assumptions identified in our filings with the Securities and Exchange Commission (the “SEC”), including our most recent Form 10-K and Form 10-Q filed with the SEC, and any subsequent filings with the SEC. Moreover, we operate in a very competitive and rapidly changing environment and it is not possible for our management to predict all risks, nor can we assess the impact of all factors on our business or the extent to which any factor, or combination of factors, may cause actual results to differ materially from those contained in any forward-looking statements we may make. In light of these risks, uncertainties and assumptions, the forward-looking statements and circumstances discussed in this presentation may not occur and actual results could differ materially and adversely from those anticipated or implied in the forward-looking statements. You should not rely upon forward-looking statements as predictions of future events. Although we believe that the expectations reflected in the forward-looking statements are reasonable, we cannot guarantee that the future results, levels of activity, performance or events and circumstances reflected in the forward-looking statements will be achieved or occur. Moreover, except as required by law, neither we nor any other person assumes responsibility for the accuracy and completeness of the forward-looking statements. We undertake no obligation to update publicly any forward-looking statements for any reason after the date of this presentation to conform these statements to actual results or to changes in our expectations, unless required by law. 2
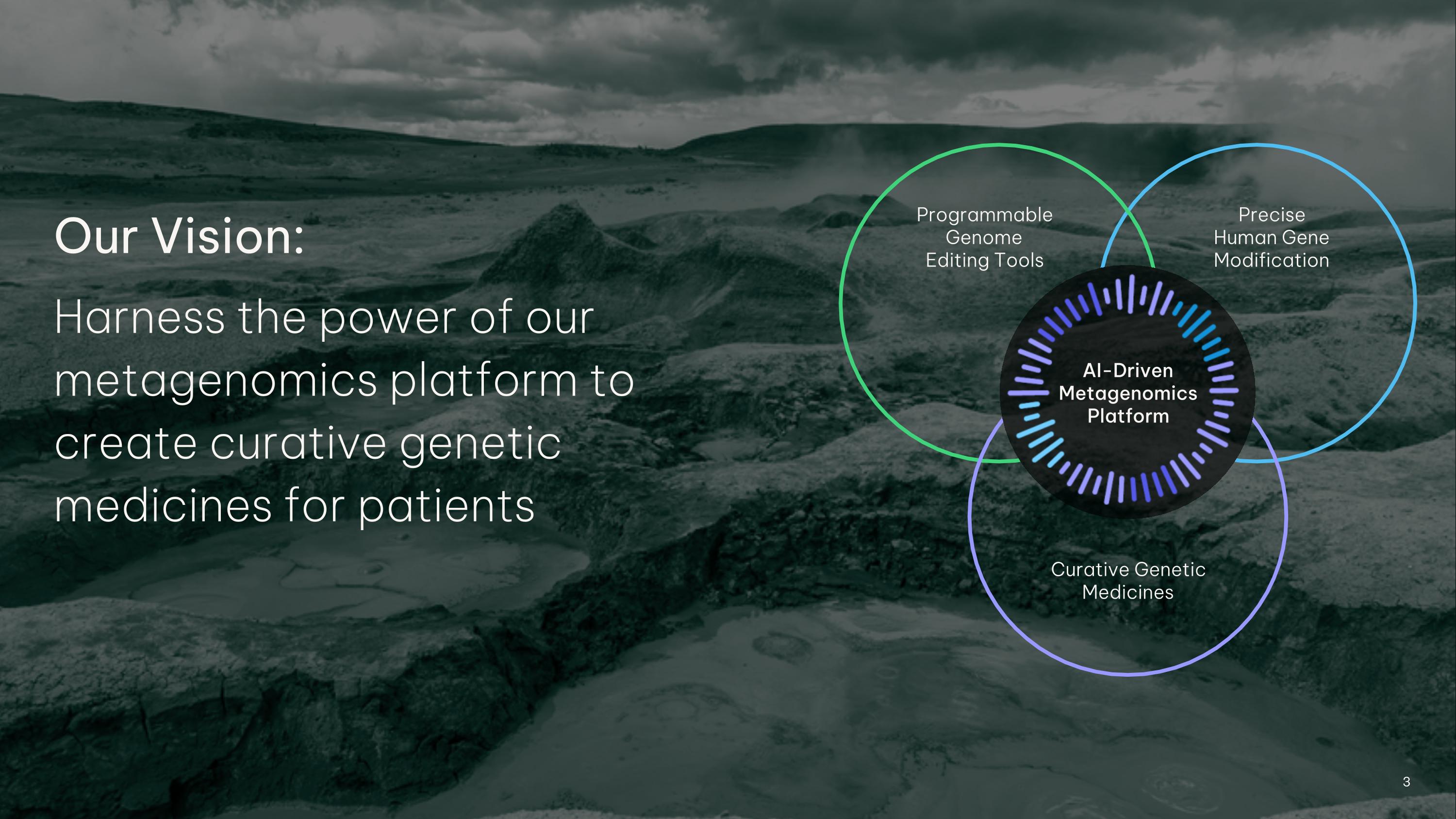
3 Our Vision: Harness the power of our metagenomics platform to create curative genetic medicines for patients Programmable Genome �Editing Tools Precise �Human Gene Modification Curative Genetic Medicines AI-Driven Metagenomics Platform
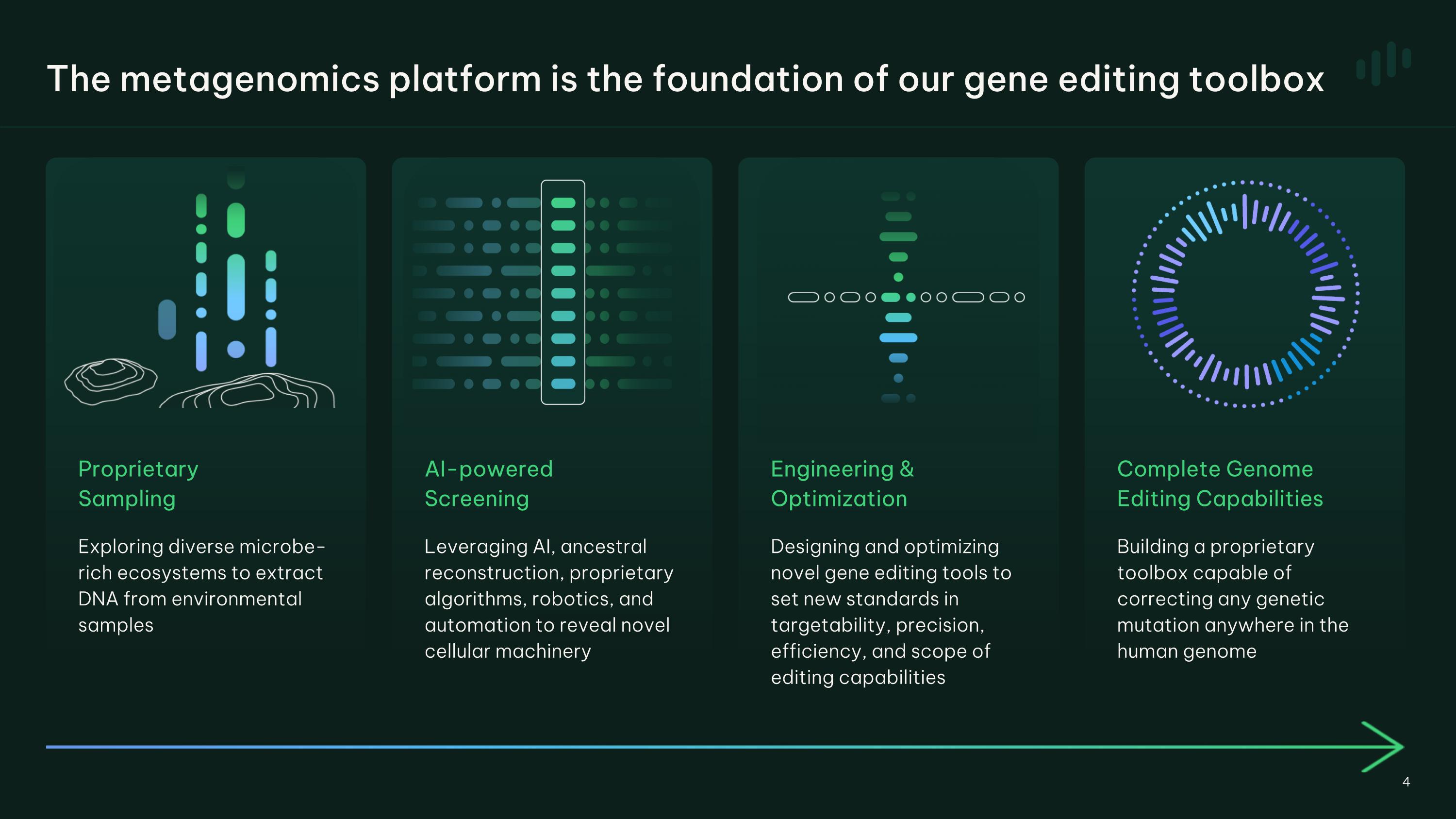
Proprietary �Sampling Exploring diverse microbe- rich ecosystems to extract DNA from environmental samples AI-powered�Screening Leveraging AI, ancestral reconstruction, proprietary algorithms, robotics, and automation to reveal novel cellular machinery Complete Genome Editing Capabilities Building a proprietary toolbox capable of correcting any genetic mutation anywhere in the human genome Engineering & Optimization Designing and optimizing novel gene editing tools to set new standards in targetability, precision, efficiency, and scope of editing capabilities The metagenomics platform is the foundation of our gene editing toolbox
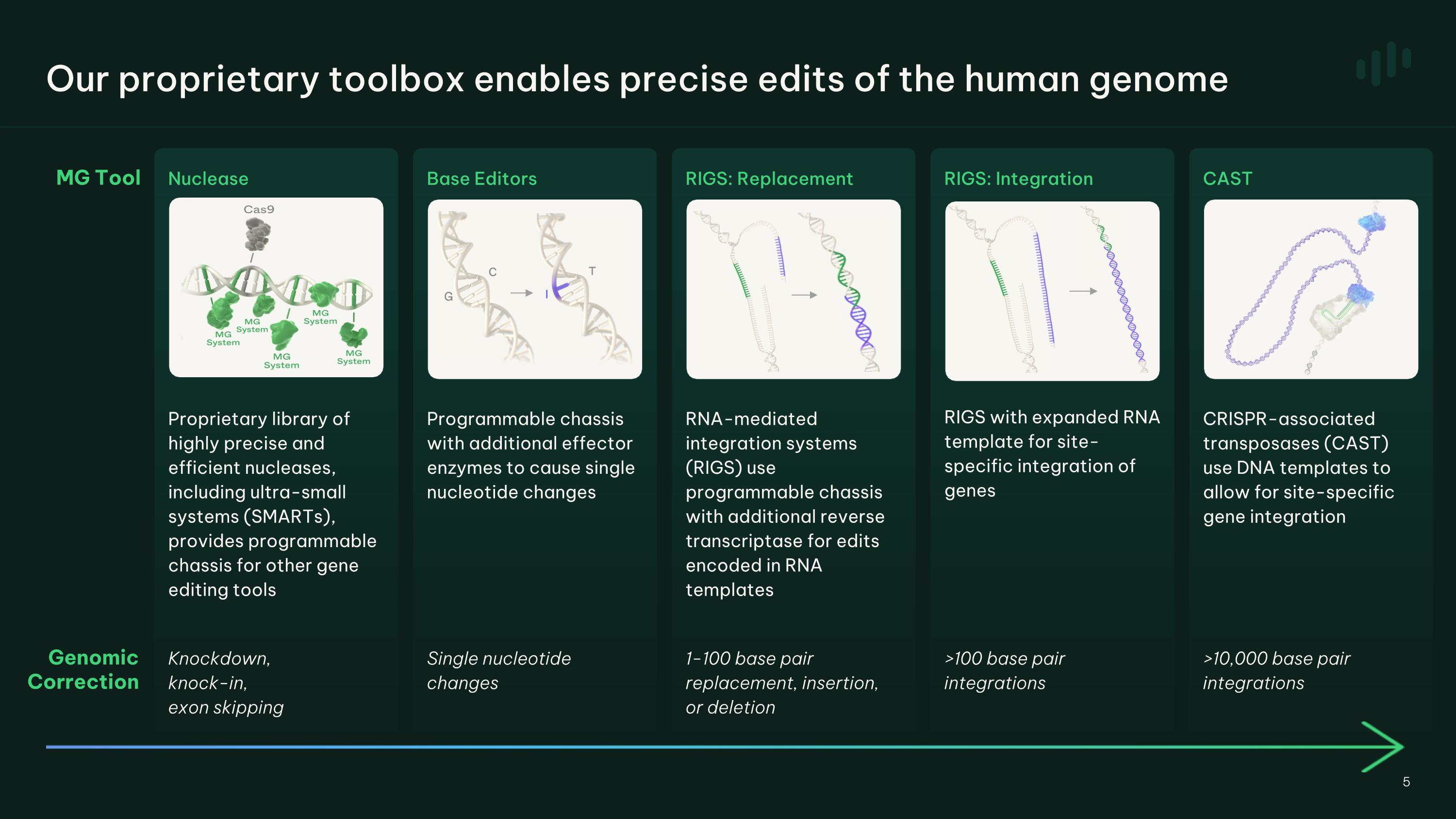
Our proprietary toolbox enables precise edits of the human genome Programmable chassis with additional effector enzymes to cause single nucleotide changes Knockdown, �knock-in, �exon skipping Single nucleotide changes Proprietary library of highly precise and efficient nucleases, including ultra-small systems (SMARTs), provides programmable chassis for other gene editing tools RNA-mediated integration systems (RIGS) use programmable chassis with additional reverse transcriptase for edits encoded in RNA templates 1-100 base pair replacement, insertion, or deletion >100 base pair integrations RIGS with expanded RNA template for site-specific integration of genes >10,000 base pair integrations CRISPR-associated transposases (CAST) use DNA templates to allow for site-specific gene integration Genomic Correction MG Tool Base Editors RIGS: Replacement RIGS: Integration CAST Nuclease 5

Editing Platform Delivery Indication / Editing Target Discovery Lead Optimization IND-Enabling Clinical LIVER LNP + AAV Hemophilia A / ALB Knock-in Undisclosed secreted protein diseases Knockdown LNP Transthyretin Amyloidosis / TTR Refractory Hypertension / AGT Undisclosed cardiovascular disease Undisclosed cardiovascular disease Small gene corrections LNP Alpha 1 Antitrypsin Deficiency / SERPINA1 Large gene insertion LNP Wilson’s Disease / ATP7B CELL�THERAPY Multiplex editing Ex vivo Solid tumor indications / TCR T Cells NEURO-�MUSCULAR Ultra small systems LNP / AAV LUNG,�KIDNEY Large gene insertion LNP / AAV 6 Partner Programs in Research: Familial ALS, Duchenne Muscular Dystrophy, Charcot Marie Tooth Disease Programs in Research: Undisclosed renal diseases, Cystic Fibrosis Multiplex editing: Undisclosed cell therapy �applications Other Program: Primary Hyperoxaluria Type 1 / HAO1 Broad pipeline built on our gene editing platform 6
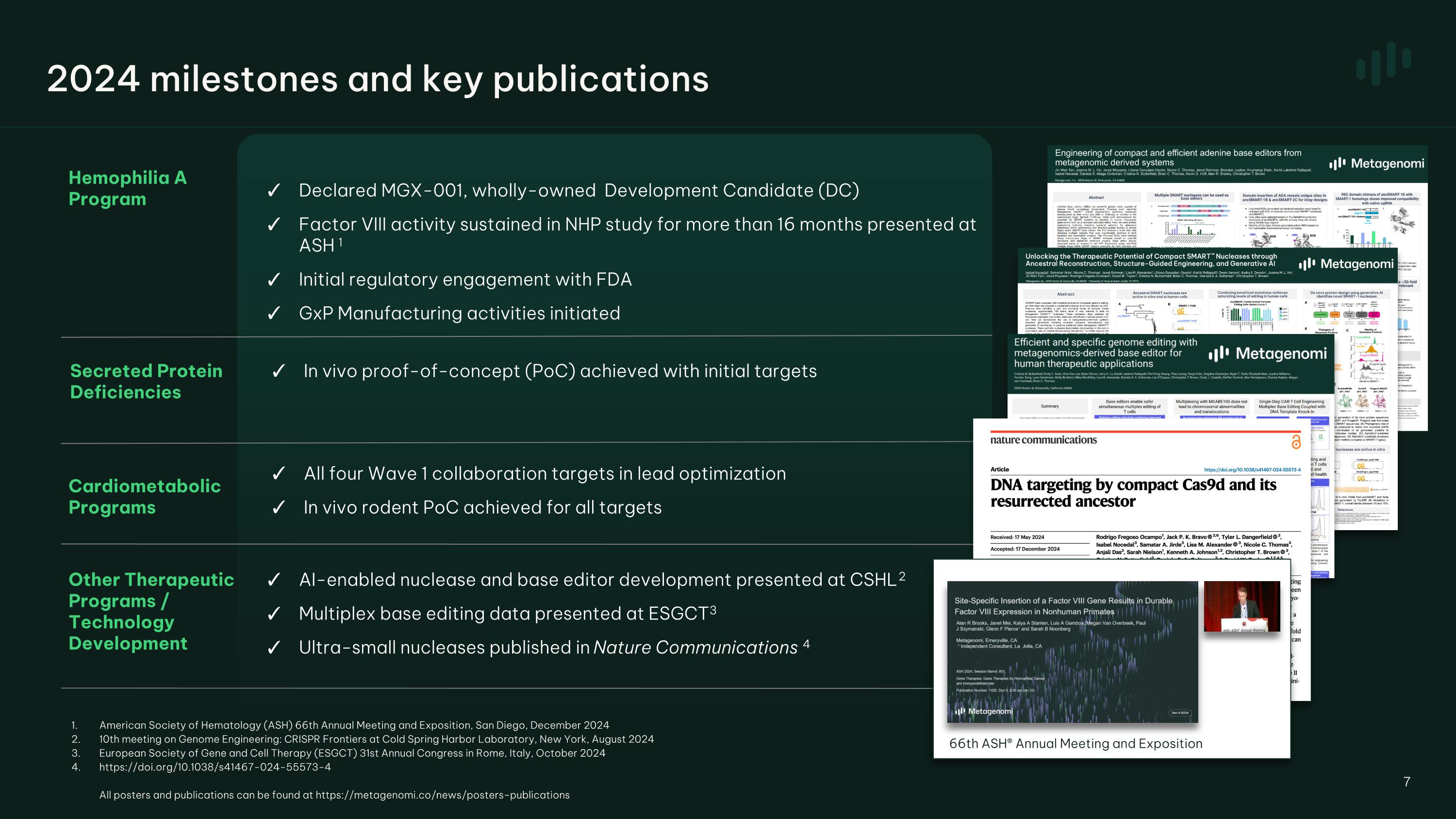
Hemophilia A Program Cardiometabolic Programs Other Therapeutic Programs / Technology Development Declared MGX-001, wholly-owned Development Candidate (DC) Factor VIII activity sustained in NHP study for more than 16 months presented at ASH 1 Initial regulatory engagement with FDA GxP Manufacturing activities initiated AI-enabled nuclease and base editor development presented at CSHL 2 Multiplex base editing data presented at ESGCT 3 Ultra-small nucleases published in Nature Communications 4 All four Wave 1 collaboration targets in lead optimization In vivo rodent PoC achieved for all targets 7 2024 milestones and key publications In vivo proof-of-concept (PoC) achieved with initial targets Secreted Protein�Deficiencies American Society of Hematology (ASH) 66th Annual Meeting and Exposition, San Diego, December 2024 10th meeting on Genome Engineering: CRISPR Frontiers at Cold Spring Harbor Laboratory, New York, August 2024 European Society of Gene and Cell Therapy (ESGCT) 31st Annual Congress in Rome, Italy, October 2024 https://doi.org/10.1038/s41467-024-55573-4 All posters and publications can be found at https://metagenomi.co/news/posters-publications 66th ASH® Annual Meeting and Exposition
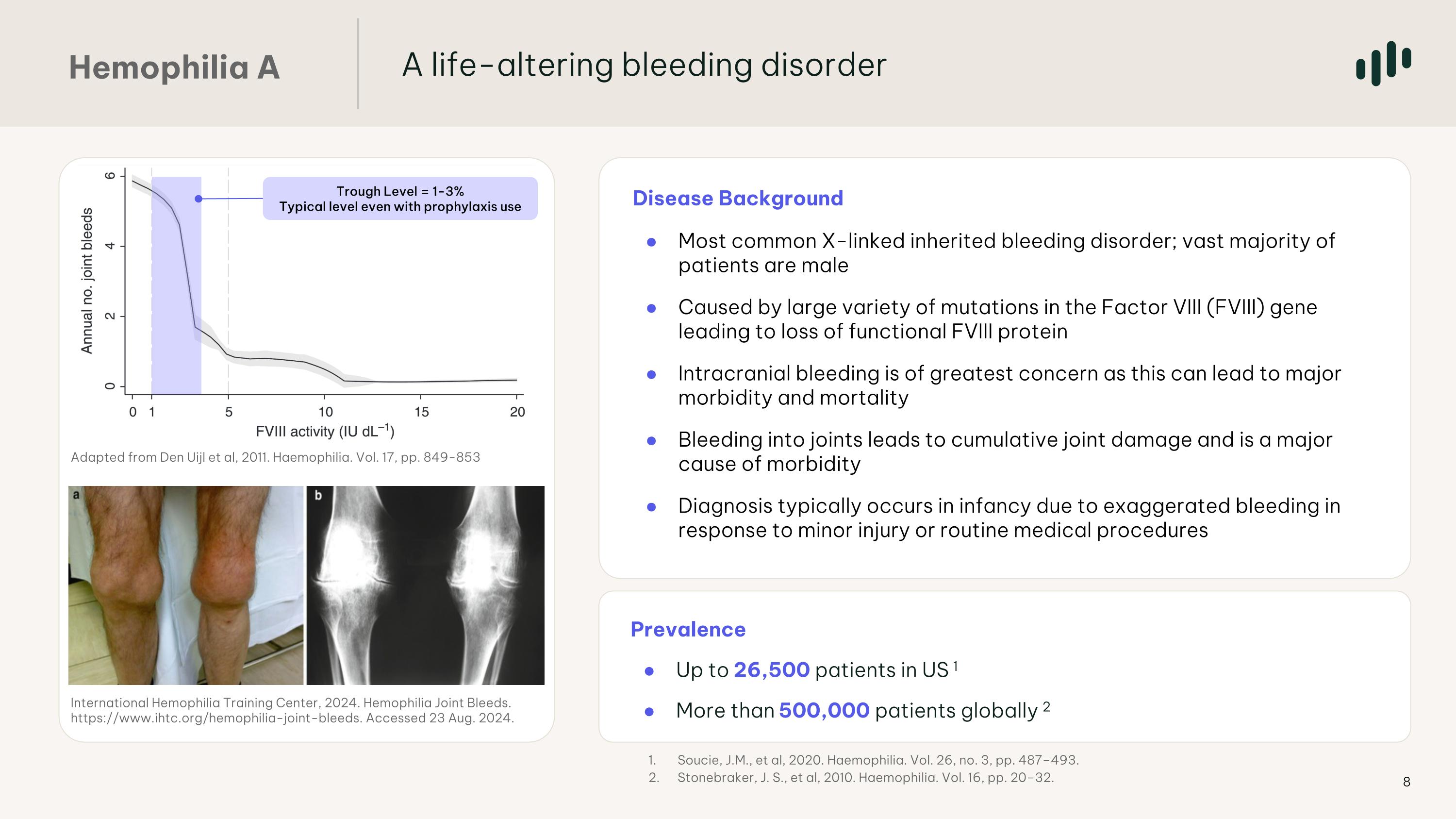
Disease Background Most common X-linked inherited bleeding disorder; vast majority of patients are male Caused by large variety of mutations in the Factor VIII (FVIII) gene leading to loss of functional FVIII protein Intracranial bleeding is of greatest concern as this can lead to major morbidity and mortality Bleeding into joints leads to cumulative joint damage and is a major cause of morbidity Diagnosis typically occurs in infancy due to exaggerated bleeding in response to minor injury or routine medical procedures Prevalence Up to 26,500 patients in US 1 More than 500,000 patients globally 2 International Hemophilia Training Center, 2024. Hemophilia Joint Bleeds. https://www.ihtc.org/hemophilia-joint-bleeds. Accessed 23 Aug. 2024. Adapted from Den Uijl et al, 2011. Haemophilia. Vol. 17, pp. 849-853 Trough Level = 1-3% Typical level even with prophylaxis use Soucie, J.M., et al, 2020. Haemophilia. Vol. 26, no. 3, pp. 487–493. Stonebraker, J. S., et al, 2010. Haemophilia. Vol. 16, pp. 20–32. A life-altering bleeding disorder Hemophilia A
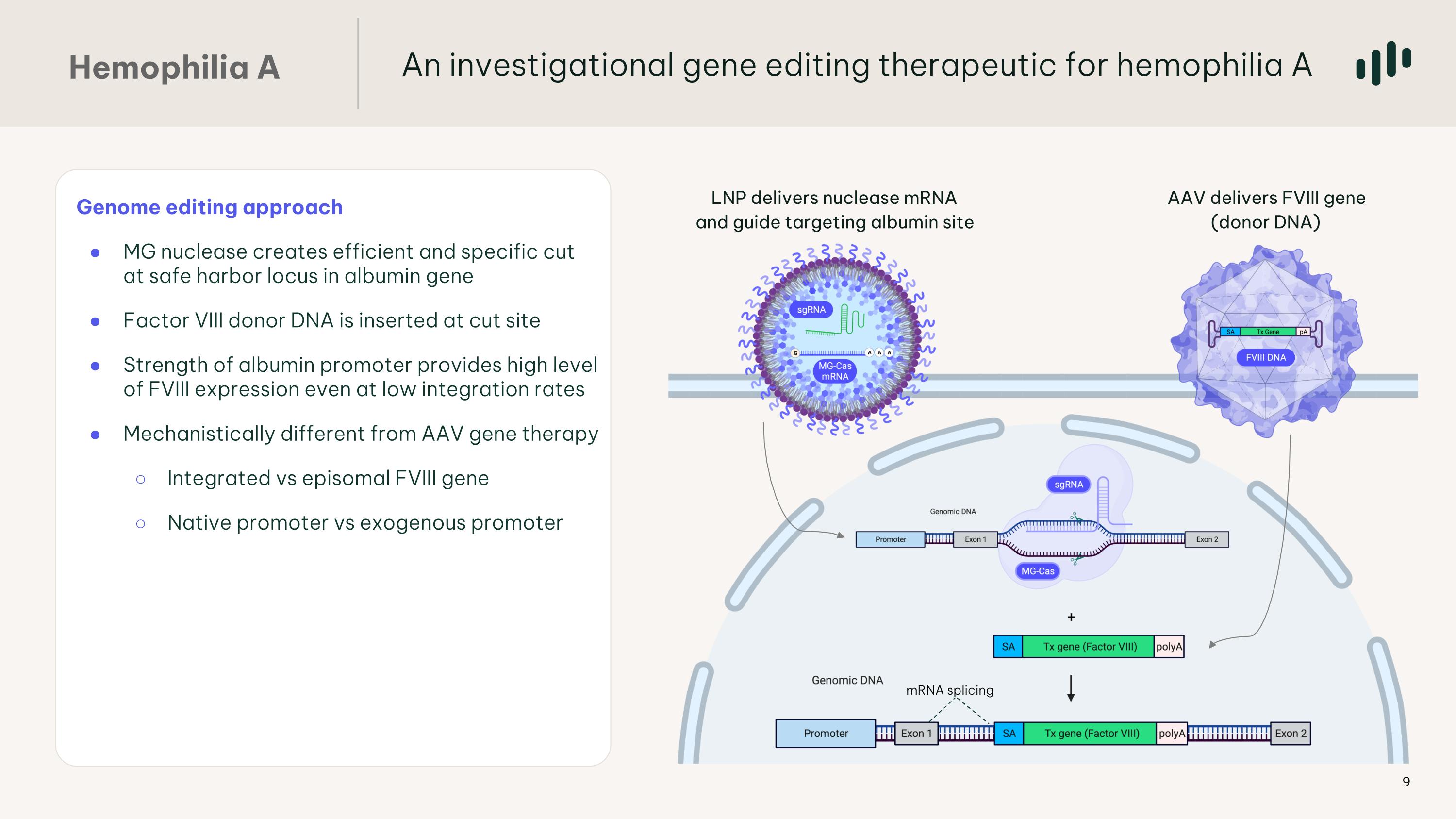
Genome editing approach MG nuclease creates efficient and specific cut at safe harbor locus in albumin gene Factor VIII donor DNA is inserted at cut site Strength of albumin promoter provides high level of FVIII expression even at low integration rates Mechanistically different from AAV gene therapy Integrated vs episomal FVIII gene Native promoter vs exogenous promoter AAV delivers FVIII gene (donor DNA) LNP delivers nuclease mRNA and guide targeting albumin site mRNA splicing An investigational gene editing therapeutic for hemophilia A Hemophilia A
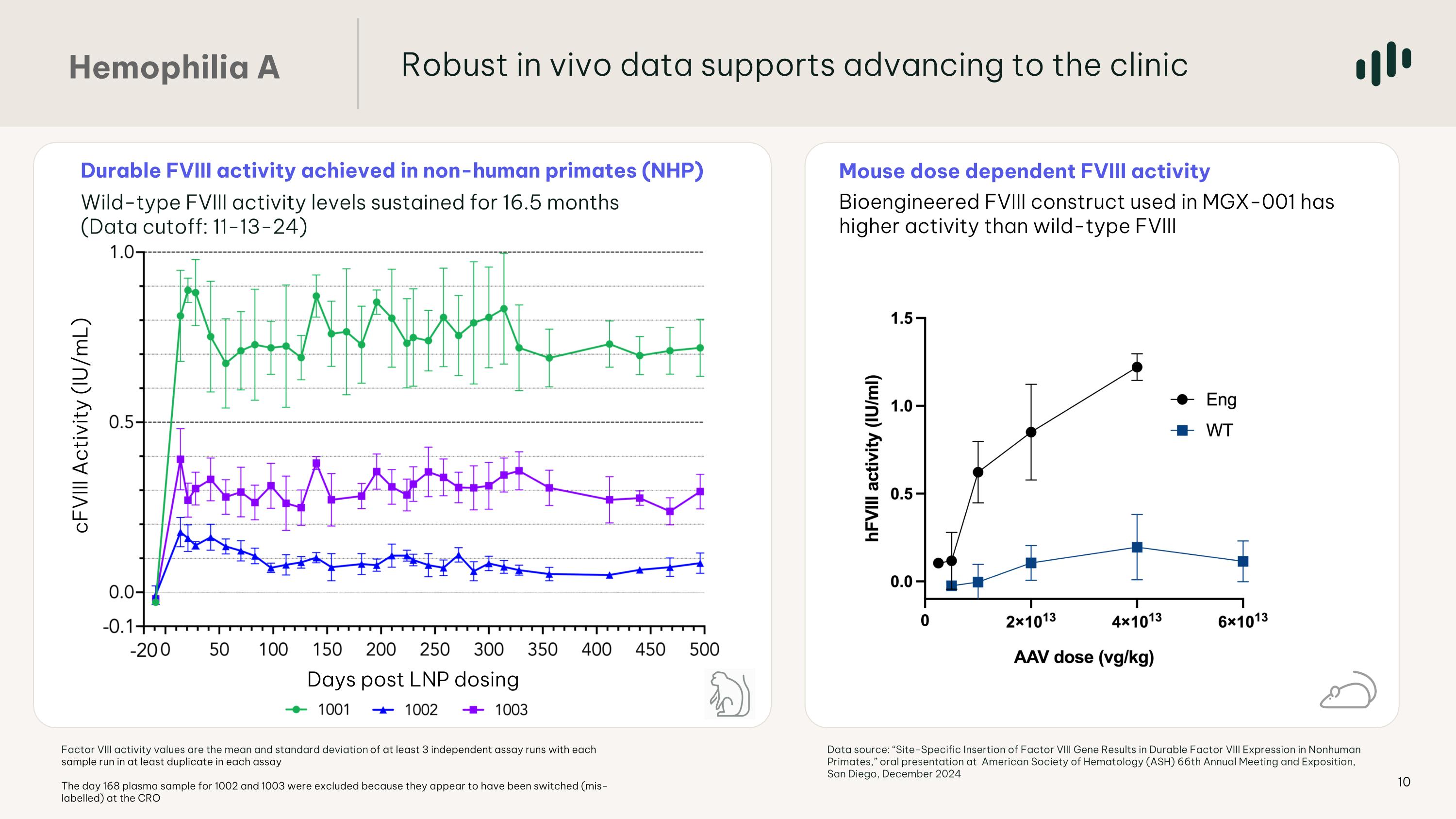
Mouse dose dependent FVIII activity Bioengineered FVIII construct used in MGX-001 has higher activity than wild-type FVIII Durable FVIII activity achieved in non-human primates (NHP) Wild-type FVIII activity levels sustained for 16.5 months (Data cutoff: 11-13-24) Days post LNP dosing cFVIII Activity (IU/mL) Hemophilia A Robust in vivo data supports advancing to the clinic Factor VIII activity values are the mean and standard deviation of at least 3 independent assay runs with each sample run in at least duplicate in each assay The day 168 plasma sample for 1002 and 1003 were excluded because they appear to have been switched (mis-labelled) at the CRO Data source: “Site-Specific Insertion of Factor VIII Gene Results in Durable Factor VIII Expression in Nonhuman Primates,” oral presentation at American Society of Hematology (ASH) 66th Annual Meeting and Exposition, San Diego, December 2024
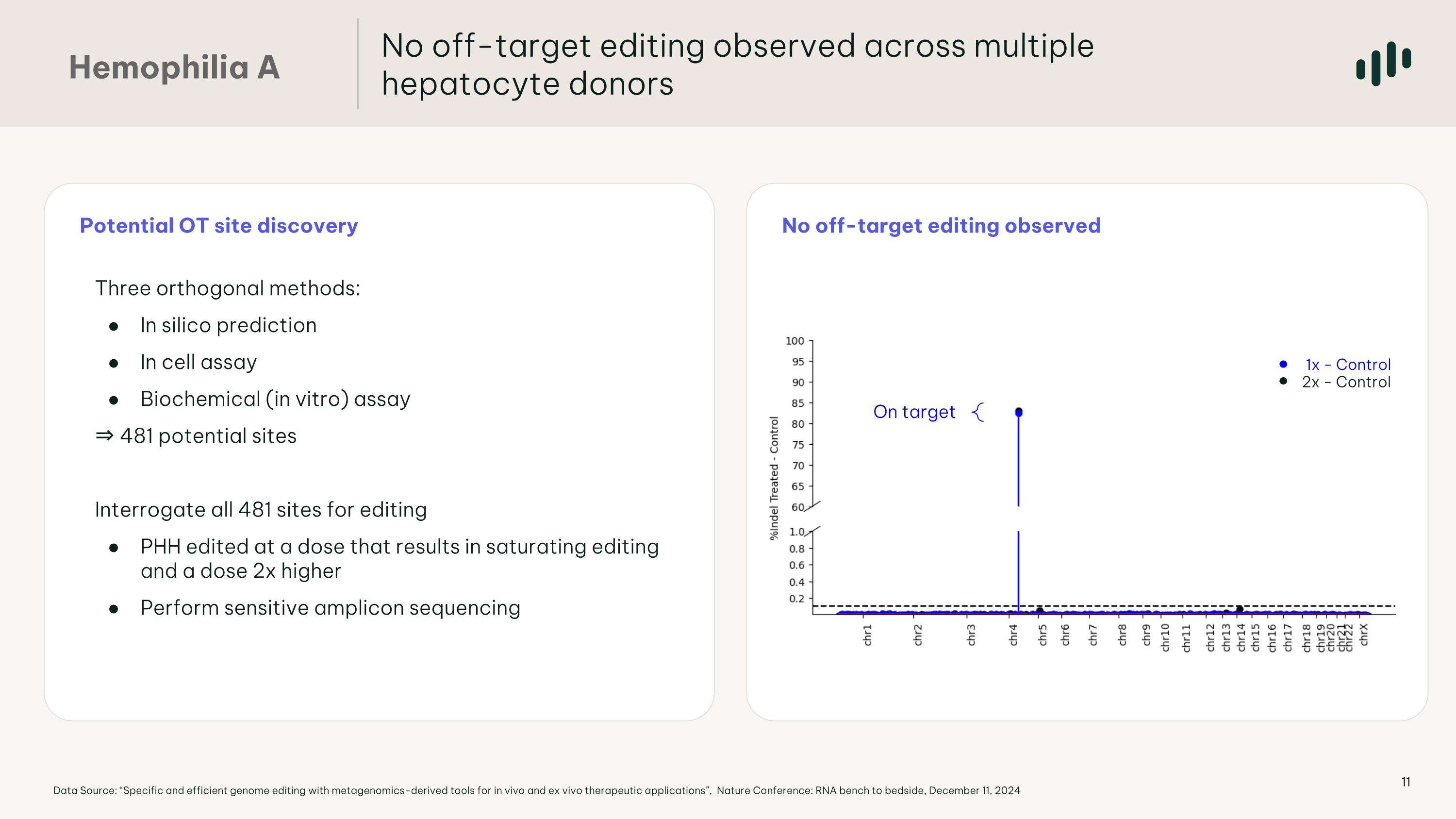
Potential OT site discovery No off-target editing observed On target 1x - Control 2x - Control No off-target editing observed across multiple hepatocyte donors Hemophilia A Data Source: “Specific and efficient genome editing with metagenomics-derived tools for in vivo and ex vivo therapeutic applications”, Nature Conference: RNA bench to bedside, December 11, 2024 Hits seen 2/5 times: 0 Hits seen 3/5 times: 0 Hits seen 4/5 times: 0 Hits seen 5/5 times: 0 Hits seen 2/5 times: 47 Hits seen 3/5 times: 7 Hits seen 4/5 times: 3 Hits seen 5/5 times: 2 Three orthogonal methods: In silico prediction In cell assay Biochemical (in vitro) assay ⇒ 481 potential sites Interrogate all 481 sites for editing PHH edited at a dose that results in saturating editing and a dose 2x higher Perform sensitive amplicon sequencing
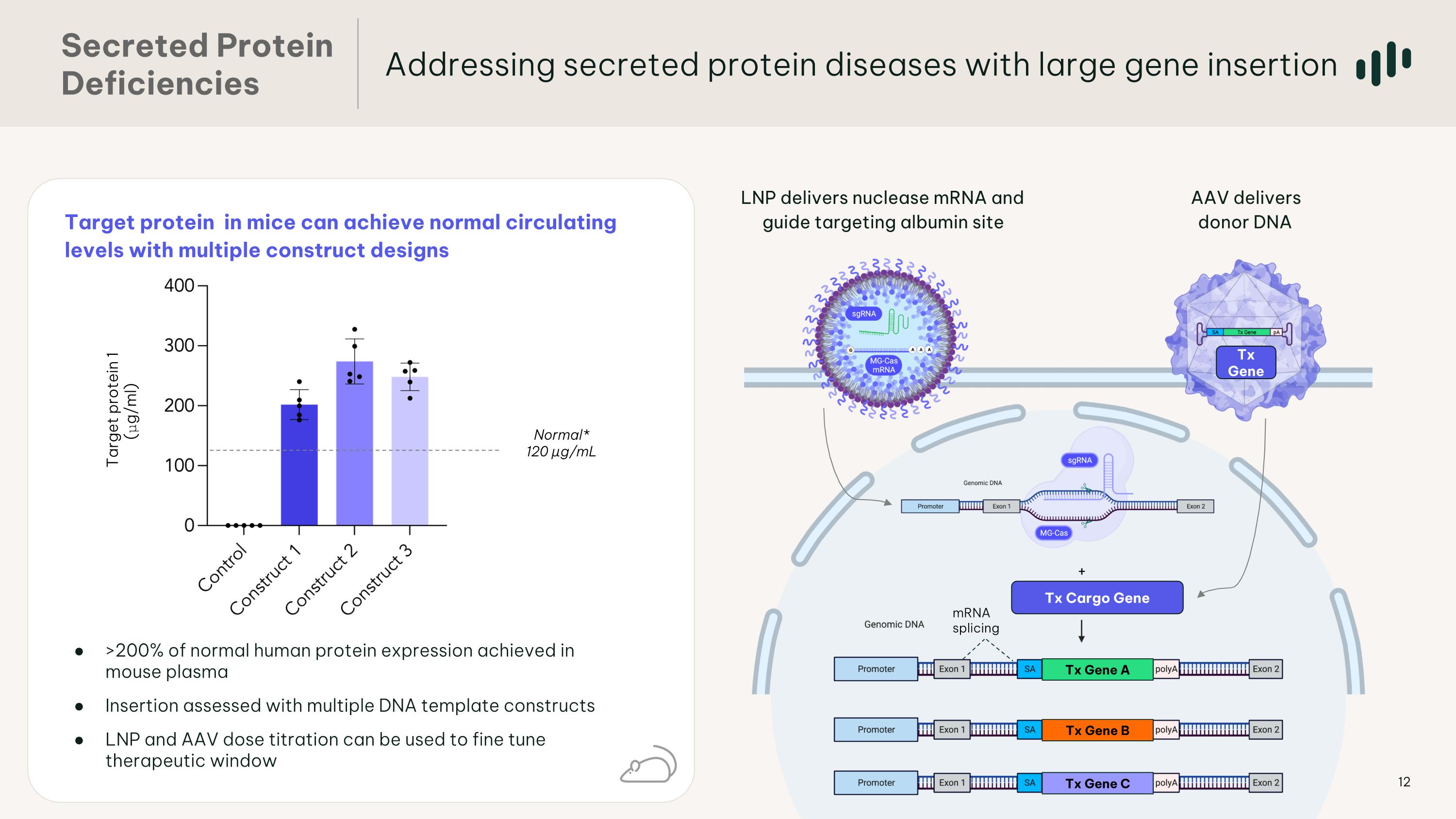
Target protein in mice can achieve normal circulating levels with multiple construct designs Addressing secreted protein diseases with large gene insertion Secreted Protein �Deficiencies AAV delivers donor DNA LNP delivers nuclease mRNA and guide targeting albumin site mRNA splicing Tx Gene Tx Gene A Tx Gene B Tx Gene C Tx Gene A Tx Cargo Gene Normal* 120 µg/mL >200% of normal human protein expression achieved in mouse plasma Insertion assessed with multiple DNA template constructs LNP and AAV dose titration can be used to fine tune therapeutic window Target protein 1 (μg/ml)
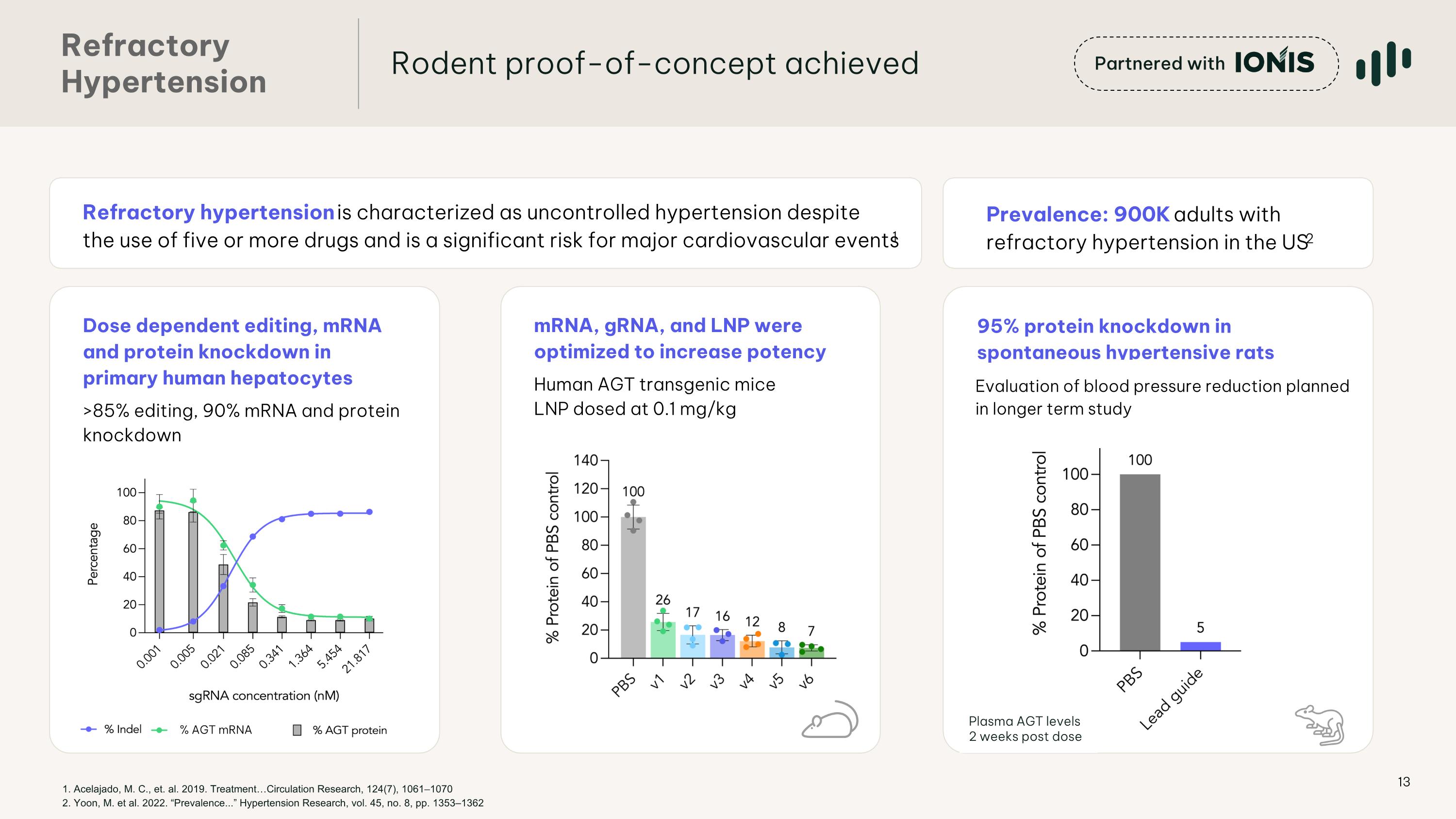
95% protein knockdown in spontaneous hypertensive rats Dose dependent editing, mRNA and protein knockdown in primary human hepatocytes >85% editing, 90% mRNA and protein knockdown Refractory �Hypertension Rodent proof-of-concept achieved Partnered with mRNA, gRNA, and LNP were optimized to increase potency Human AGT transgenic mice LNP dosed at 0.1 mg/kg % AGT mRNA Evaluation of blood pressure reduction planned in longer term study Plasma AGT levels 2 weeks post dose 1. Acelajado, M. C., et. al. 2019. Treatment…Circulation Research, 124(7), 1061–1070�2. Yoon, M. et al. 2022. “Prevalence...” Hypertension Research, vol. 45, no. 8, pp. 1353–1362 Prevalence: 900K adults with refractory hypertension in the US2 Refractory hypertension is characterized as uncontrolled hypertension despite the use of five or more drugs and is a significant risk for major cardiovascular events1
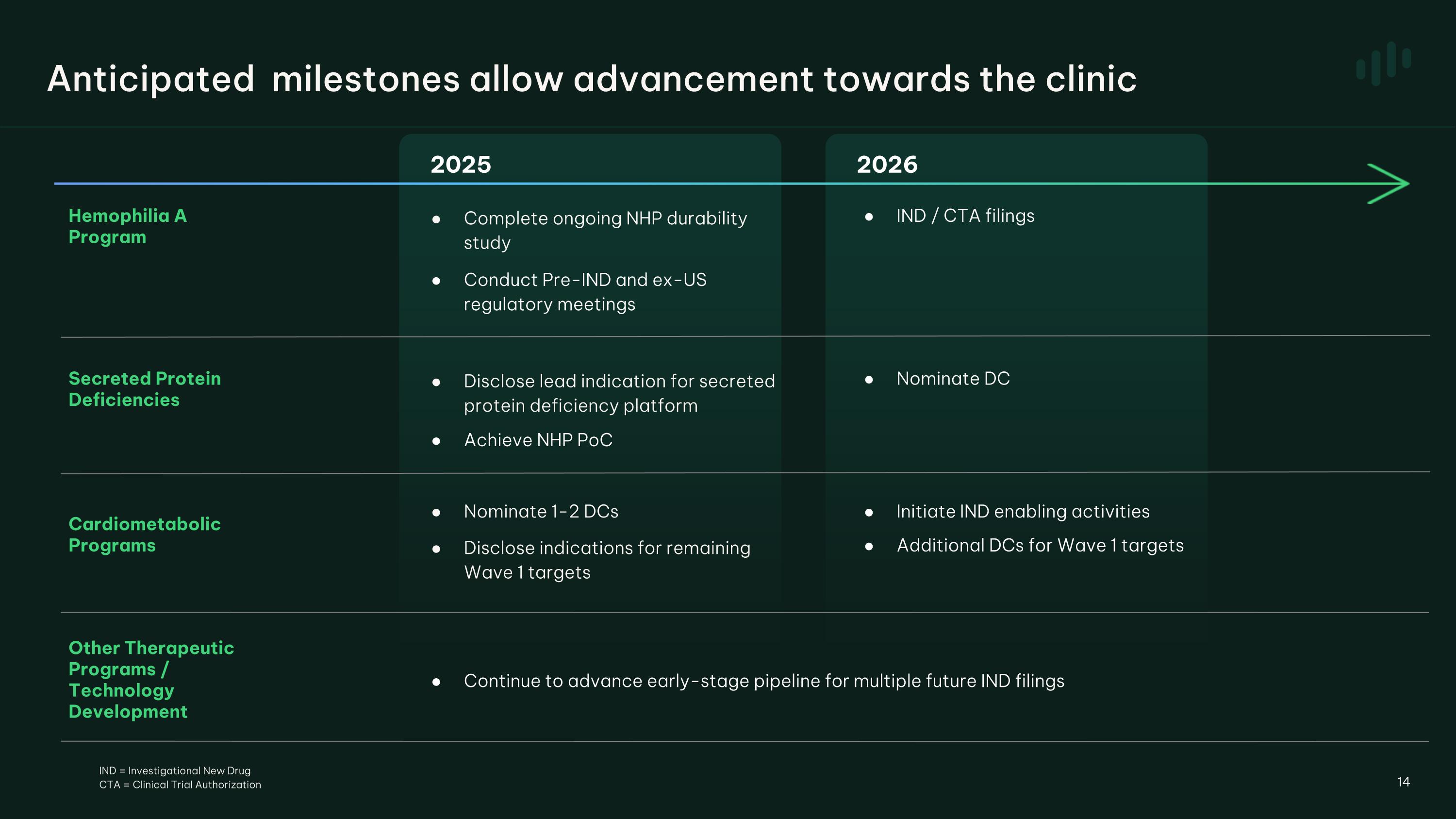
2025 2026 Hemophilia A Program Complete ongoing NHP durability study Conduct Pre-IND and ex-US regulatory meetings IND / CTA filings Cardiometabolic Programs Other Therapeutic Programs / Technology Development Continue to advance early-stage pipeline for multiple future IND filings Nominate 1-2 DCs Disclose indications for remaining Wave 1 targets Initiate IND enabling activities Additional DCs for Wave 1 targets 14 Anticipated milestones allow advancement towards the clinic Nominate DC Disclose lead indication for secreted protein deficiency platform Achieve NHP PoC Secreted Protein�Deficiencies IND = Investigational New Drug CTA = Clinical Trial Authorization
Metagenomi: Pipeline advancing, positioned for success Well capitalized with cash runway into 2027 Harnessing the power of our metagenomics platform to create curative genetic medicines for patients Advancing cardiometabolic programs with Ionis Progressing additional wholly owned therapeutic candidates Driving towards clinic with wholly-owned MGX-001 in hemophilia A Leveraging MGX-001 to establish secreted protein deficiencies platform Realizing the potential of our AI-enabled gene editing capabilities 15














